Activation of multiple signaling pathways causes developmental defects in mice with a Noonan syndrome–associated Sos1 mutation
- PMID: 21041952
- PMCID: PMC2993597
- DOI: 10.1172/JCI43910
Activation of multiple signaling pathways causes developmental defects in mice with a Noonan syndrome–associated Sos1 mutation
Abstract
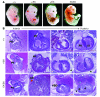
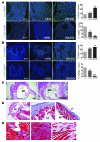
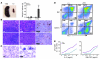
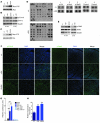
Noonan syndrome (NS) is an autosomal dominant genetic disorder characterized by short stature, unique facial features, and congenital heart disease. About 10%-15% of individuals with NS have mutations in son of sevenless 1 (SOS1), which encodes a RAS and RAC guanine nucleotide exchange factor (GEF). To understand the role of SOS1 in the pathogenesis of NS, we generated mice with the NS-associated Sos1E846K gain-of-function mutation. Both heterozygous and homozygous mutant mice showed many NS-associated pheno-types, including growth delay, distinctive facial dysmorphia, hematologic abnormalities, and cardiac defects. We found that the Ras/MAPK pathway as well as Rac and Stat3 were activated in the mutant hearts. These data provide in vivo molecular and cellular evidence that Sos1 is a GEF for Rac under physiological conditions and suggest that Rac and Stat3 activation might contribute to NS phenotypes. Furthermore, prenatal administration of a MEK inhibitor ameliorated the embryonic lethality, cardiac defects, and NS features of the homozygous mutant mice, demonstrating that this signaling pathway might represent a promising therapeutic target for NS.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

