Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa
- PMID: 21042406
- PMCID: PMC2958839
- DOI: 10.1371/journal.pone.0013557
Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa
Abstract
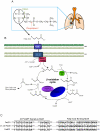
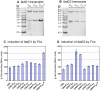
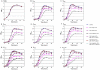
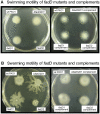
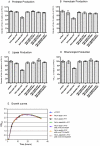
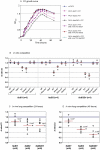
A close interconnection between nutrient metabolism and virulence factor expression contributes to the pathophysiology of Pseudomonas aeruginosa as a successful pathogen. P. aeruginosa fatty acid (FA) degradation is complicated with multiple acyl-CoA synthetase homologs (FadDs) expressed in vivo in lung tissue during cystic fibrosis infections. The promoters of two genetically linked P. aeruginosa fadD genes (fadD1 and fadD2) were mapped and northern blot analysis indicated they could exist on two different transcripts. These FadDs contain ATP/AMP signature and FA-binding motifs highly homologous to those of the Escherichia coli FadD. Upon introduction into an E. coli fadD(-)/fadR(-) double mutant, both P. aeruginosa fadDs functionally complemented the E. coli fadD(-)/fadR(-) mutant, allowing degradation of different chain-length FAs. Chromosomal mutagenesis, growth analysis, induction studies, and determination of kinetic parameters suggested that FadD1 has a substrate preference for long-chain FAs while FadD2 prefers shorter-chain FAs. When compared to the wild type strain, the fadD2 mutant exhibited decreased production of lipase, protease, rhamnolipid and phospholipase, and retardation of both swimming and swarming motilities. Interestingly, fadD1 mutant showed only increased swarming motility. Growth analysis of the fadD mutants showed noticeable deficiencies in utilizing FAs and phosphatidylcholine (major components of lung surfactant) as the sole carbon source. This defect translated into decreased in vivo fitness of P. aeruginosa in a BALB/c mouse lung infection model, supporting the role of lipids as a significant nutrient source for this bacterium in vivo.
Conflict of interest statement
Figures


References
-
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. - PubMed
-
- Abd H, Wretlind B, Saeed A, Idsund E, Hultenby K, et al. Pseudomonas aeruginosa utilizes its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J Eukaryot Microbiol. 2008;55:235–243. - PubMed
-
- Baltch AL, Griffin PE. Pseudomonas aeruginosa bacteremia: clinical study of 75 patients. Am J Med Sci. 1977;274:119–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

