A dyslexia-associated variant in DCDC2 changes gene expression
- PMID: 21042874
- PMCID: PMC3053575
- DOI: 10.1007/s10519-010-9408-3
A dyslexia-associated variant in DCDC2 changes gene expression
Abstract
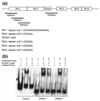
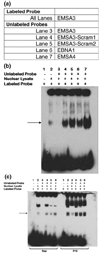
Reading disability (RD) or dyslexia is a common neurogenetic disorder. Two genes, KIAA0319 and DCDC2, have been identified by association studies of the DYX2 locus on 6p21.3. We previously identified a 2445 bp deletion, and a compound STR within the deleted region (BV677278), in intron 2 of DCDC2. The deletion and several alleles of the STR are strongly associated with RD (P = 0.00002). In this study we investigated whether BV677278 is a regulatory region for DCDC2 by electrophoretic mobility shift and luciferase reporter assays. We show that oligonucleotide probes from the STR bind nuclear protein from human brain, and that alleles of the STR have a range of DCDC2-specific enhancer activities. Five alleles displayed strong enhancer activity and increased gene expression, while allele 1 showed no enhancer activity. These studies suggest that the association of BV677278 with RD reflects a role as a modifier of DCDC2 expression.
Figures

References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300.
-
- Brkanac Z, Chapman NH, Matsushita MM, Chun L, Nielsen K, Cochrane E, et al. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(4):556–560. - PubMed
-
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266(5183):276–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

