BLV-CoCoMo-qPCR: Quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm
- PMID: 21044304
- PMCID: PMC2988707
- DOI: 10.1186/1742-4690-7-91
BLV-CoCoMo-qPCR: Quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm
Abstract
Background: Bovine leukemia virus (BLV) is closely related to human T-cell leukemia virus (HTLV) and is the etiological agent of enzootic bovine leukosis, a disease characterized by a highly extended course that often involves persistent lymphocytosis and culminates in B-cell lymphomas. BLV provirus remains integrated in cellular genomes, even in the absence of detectable BLV antibodies. Therefore, to understand the mechanism of BLV-induced leukemogenesis and carry out the selection of BLV-infected animals, a detailed evaluation of changes in proviral load throughout the course of disease in BLV-infected cattle is required. The aim of this study was to develop a new quantitative real-time polymerase chain reaction (PCR) method using Coordination of Common Motifs (CoCoMo) primers to measure the proviral load of known and novel BLV variants in clinical animals.
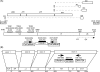
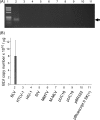
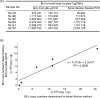
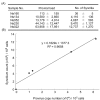
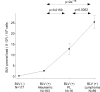
Results: Degenerate primers were designed from 52 individual BLV long terminal repeat (LTR) sequences identified from 356 BLV sequences in GenBank using the CoCoMo algorithm, which has been developed specifically for the detection of multiple virus species. Among 72 primer sets from 49 candidate primers, the most specific primer set was selected for detection of BLV LTR by melting curve analysis after real-time PCR amplification. An internal BLV TaqMan probe was used to enhance the specificity and sensitivity of the assay, and a parallel amplification of a single-copy host gene (the bovine leukocyte antigen DRA gene) was used to normalize genomic DNA. The assay is highly specific, sensitive, quantitative and reproducible, and was able to detect BLV in a number of samples that were negative using the previously developed nested PCR assay. The assay was also highly effective in detecting BLV in cattle from a range of international locations. Finally, this assay enabled us to demonstrate that proviral load correlates not only with BLV infection capacity as assessed by syncytium formation, but also with BLV disease progression.
Conclusions: Using our newly developed BLV-CoCoMo-qPCR assay, we were able to detect a wide range of mutated BLV viruses. CoCoMo algorithm may be a useful tool to design degenerate primers for quantification of proviral load for other retroviruses including HTLV and human immunodeficiency virus type 1.
Figures




Similar articles
-
BLV-CoCoMo-qPCR: a useful tool for evaluating bovine leukemia virus infection status.BMC Vet Res. 2012 Sep 21;8:167. doi: 10.1186/1746-6148-8-167. BMC Vet Res. 2012. PMID: 22995575 Free PMC article.
-
BLV-CoCoMo-qPCR-2: improvements to the BLV-CoCoMo-qPCR assay for bovine leukemia virus by reducing primer degeneracy and constructing an optimal standard curve.Arch Virol. 2015 May;160(5):1325-32. doi: 10.1007/s00705-015-2377-3. Epub 2015 Mar 4. Arch Virol. 2015. PMID: 25731158
-
Risk factors associated with increased bovine leukemia virus proviral load in infected cattle in Japan from 2012 to 2014.Virus Res. 2015 Dec 2;210:283-90. doi: 10.1016/j.virusres.2015.08.020. Epub 2015 Aug 29. Virus Res. 2015. PMID: 26321160
-
Viral oncogenesis of δ-retroviruses, HTLV-1 and BLV, and recent advances in its diagnosis.Virology. 2025 Apr;605:110461. doi: 10.1016/j.virol.2025.110461. Epub 2025 Feb 21. Virology. 2025. PMID: 40015031 Review.
-
Epidemiology and genetic diversity of bovine leukemia virus.Virol J. 2017 Nov 2;14(1):209. doi: 10.1186/s12985-017-0876-4. Virol J. 2017. PMID: 29096657 Free PMC article. Review.
Cited by
-
Encephalomyocarditis virus is potentially derived from eastern bent-wing bats living in East Asian countries.Virus Res. 2019 Jan 2;259:62-67. doi: 10.1016/j.virusres.2018.10.020. Epub 2018 Nov 1. Virus Res. 2019. PMID: 30391400 Free PMC article.
-
Development of a predictive model for bovine leukemia virus proviral load.J Vet Intern Med. 2022 Sep;36(5):1827-1836. doi: 10.1111/jvim.16506. Epub 2022 Aug 11. J Vet Intern Med. 2022. PMID: 35950569 Free PMC article.
-
Visualizing bovine leukemia virus (BLV)-infected cells and measuring BLV proviral loads in the milk of BLV seropositive dams.Vet Res. 2019 Nov 29;50(1):102. doi: 10.1186/s13567-019-0724-1. Vet Res. 2019. PMID: 31783914 Free PMC article.
-
Three YXXL Sequences of a Bovine Leukemia Virus Transmembrane Protein are Independently Required for Fusion Activity by Controlling Expression on the Cell Membrane.Viruses. 2019 Dec 10;11(12):1140. doi: 10.3390/v11121140. Viruses. 2019. PMID: 31835517 Free PMC article.
-
Comparative Evaluation of Three Commercial Quantitative Real-Time PCRs Used in Japan for Bovine Leukemia Virus.Viruses. 2022 May 28;14(6):1182. doi: 10.3390/v14061182. Viruses. 2022. PMID: 35746654 Free PMC article.
References
-
- Endoh D, Mizutani T, Kirisawa R, Maki Y, Saito H, Kon Y, Morikawa S, Hayashi M. Species-independent detection of RNA virus by representational difference analysis using non-ribosomal hexanucleotides for reverse transcription. Nucleic Acids Res. 2005;33:e65. doi: 10.1093/nar/gni064. - DOI - PMC - PubMed
-
- Bartl S. Amplification using degenerate primers with multiple inosines to isolate genes with minimal sequence similarity. Methods Mol Biol. 1997;67:451–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

