Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6
- PMID: 21044320
- PMCID: PMC2987987
- DOI: 10.1186/1471-2180-10-274
Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6
Abstract
Background: Pandemic Vibrio parahaemolyticus has undergone rapid changes in both K- and O-antigens, making detection of outbreaks more difficult. In order to understand these rapid changes, the genetic regions encoding these antigens must be examined. In Vibrio cholerae and Vibrio vulnificus, both O-antigen and capsular polysaccharides are encoded in a single region on the large chromosome; a similar arrangement in pandemic V. parahaemolyticus would help explain the rapid serotype changes. However, previous reports on "capsule" genes are controversial. Therefore, we set out to clarify and characterize these regions in pandemic V. parahaemolyticus O3:K6 by gene deletion using a chitin based transformation strategy.
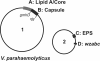
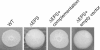
Results: We generated different deletion mutants of putative polysaccharide genes and examined the mutants by immuno-blots with O and K specific antisera. Our results showed that O- and K-antigen genes are separated in V. parahaemolyticus O3:K6; the region encoding both O-antigen and capsule biosynthesis in other vibrios, i.e. genes between gmhD and rjg, determines the K6-antigen but not the O3-antigen in V. parahaemolyticus. The previously identified "capsule genes" on the smaller chromosome were related to exopolysaccharide synthesis, not K-antigen.
Conclusion: Understanding of the genetic basis of O- and K-antigens is critical to understanding the rapid changes in these polysaccharides seen in pandemic V. parahaemolyticus. This report confirms the genetic location of K-antigen synthesis in V. parahaemolyticus O3:K6 allowing us to focus future studies of the evolution of serotypes to this region.
Figures




References
-
- Fujino L, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, Uebo T. On the bacteriological examination of shirasu food poisoning. Med J Osaka Univ. 1953;4:299–304.
-
- Nair GB, Hormazabal JC. The Vibrio parahaemolyticus pandemic. Rev Chilena Infectol. 2005;22(2):125–130. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

