Mapping transcription mechanisms from multimodal genomic data
- PMID: 21044360
- PMCID: PMC2967743
- DOI: 10.1186/1471-2105-11-S9-S2
Mapping transcription mechanisms from multimodal genomic data
Abstract
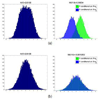
Background: Identification of expression quantitative trait loci (eQTLs) is an emerging area in genomic study. The task requires an integrated analysis of genome-wide single nucleotide polymorphism (SNP) data and gene expression data, raising a new computational challenge due to the tremendous size of data.
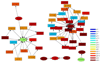
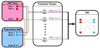
Results: We develop a method to identify eQTLs. The method represents eQTLs as information flux between genetic variants and transcripts. We use information theory to simultaneously interrogate SNP and gene expression data, resulting in a Transcriptional Information Map (TIM) which captures the network of transcriptional information that links genetic variations, gene expression and regulatory mechanisms. These maps are able to identify both cis- and trans- regulating eQTLs. The application on a dataset of leukemia patients identifies eQTLs in the regions of the GART, PCP4, DSCAM, and RIPK4 genes that regulate ADAMTS1, a known leukemia correlate.
Conclusions: The information theory approach presented in this paper is able to infer the dependence networks between SNPs and transcripts, which in turn can identify cis- and trans-eQTLs. The application of our method to the leukemia study explains how genetic variants and gene expression are linked to leukemia.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

