Using gene co-expression network analysis to predict biomarkers for chronic lymphocytic leukemia
- PMID: 21044363
- PMCID: PMC2967746
- DOI: 10.1186/1471-2105-11-S9-S5
Using gene co-expression network analysis to predict biomarkers for chronic lymphocytic leukemia
Abstract
Background: Chronic lymphocytic leukemia (CLL) is the most common adult leukemia. It is a highly heterogeneous disease, and can be divided roughly into indolent and progressive stages based on classic clinical markers. Immunoglobin heavy chain variable region (IgVH) mutational status was found to be associated with patient survival outcome, and biomarkers linked to the IgVH status has been a focus in the CLL prognosis research field. However, biomarkers highly correlated with IgVH mutational status which can accurately predict the survival outcome are yet to be discovered.
Results: In this paper, we investigate the use of gene co-expression network analysis to identify potential biomarkers for CLL. Specifically we focused on the co-expression network involving ZAP70, a well characterized biomarker for CLL. We selected 23 microarray datasets corresponding to multiple types of cancer from the Gene Expression Omnibus (GEO) and used the frequent network mining algorithm CODENSE to identify highly connected gene co-expression networks spanning the entire genome, then evaluated the genes in the co-expression network in which ZAP70 is involved. We then applied a set of feature selection methods to further select genes which are capable of predicting IgVH mutation status from the ZAP70 co-expression network.
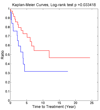
Conclusions: We have identified a set of genes that are potential CLL prognostic biomarkers IL2RB, CD8A, CD247, LAG3 and KLRK1, which can predict CLL patient IgVH mutational status with high accuracies. Their prognostic capabilities were cross-validated by applying these biomarker candidates to classify patients into different outcome groups using a CLL microarray datasets with clinical information.
Figures



Similar articles
-
ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile.Blood. 2003 Jun 15;101(12):4944-51. doi: 10.1182/blood-2002-10-3306. Epub 2003 Feb 20. Blood. 2003. PMID: 12595313
-
Regulatory network reconstruction reveals genes with prognostic value for chronic lymphocytic leukemia.BMC Genomics. 2015 Nov 25;16:1002. doi: 10.1186/s12864-015-2189-6. BMC Genomics. 2015. PMID: 26606983 Free PMC article.
-
ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia.N Engl J Med. 2003 May 1;348(18):1764-75. doi: 10.1056/NEJMoa023143. N Engl J Med. 2003. PMID: 12724482
-
[Research progress on prognostic markers of chronic lymphocytic leukemia].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2010 May;39(3):250-6. doi: 10.3785/j.issn.1008-9292.2010.03.007. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2010. PMID: 20544986 Review. Chinese.
-
[ZAP-70 in B-cell chronic lymphocytic leukemia: clinical significance and methods of detection].Vnitr Lek. 2006 Dec;52(12):1194-9. Vnitr Lek. 2006. PMID: 17299914 Review. Czech.
Cited by
-
The effects of graded levels of calorie restriction: VII. Topological rearrangement of hypothalamic aging networks.Aging (Albany NY). 2016 May;8(5):917-32. doi: 10.18632/aging.100944. Aging (Albany NY). 2016. PMID: 27115072 Free PMC article.
-
Novel immunologic approaches in lymphoma: unleashing the brakes on the immune system.Curr Oncol Rep. 2015 Jul;17(7):30. doi: 10.1007/s11912-015-0456-y. Curr Oncol Rep. 2015. PMID: 25986722 Review.
-
Pathway-based analysis of the hidden genetic heterogeneities in cancers.Genomics Proteomics Bioinformatics. 2014 Feb;12(1):31-8. doi: 10.1016/j.gpb.2013.12.001. Epub 2014 Jan 22. Genomics Proteomics Bioinformatics. 2014. PMID: 24462714 Free PMC article.
-
Rat hepatocytes weighted gene co-expression network analysis identifies specific modules and hub genes related to liver regeneration after partial hepatectomy.PLoS One. 2014 Apr 17;9(4):e94868. doi: 10.1371/journal.pone.0094868. eCollection 2014. PLoS One. 2014. PMID: 24743545 Free PMC article.
-
When is hub gene selection better than standard meta-analysis?PLoS One. 2013 Apr 17;8(4):e61505. doi: 10.1371/journal.pone.0061505. Print 2013. PLoS One. 2013. PMID: 23613865 Free PMC article.
References
-
- Shanafelt TD, Byrd JC, Call TG, Zent CS, Kay NE. Narrative review: initial management of newly diagnosed, early-stage chronic lymphocytic leukemia. Ann Intern Med. 2006;145(6):435–447. - PubMed
-
- Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46(2):219–234. - PubMed
-
- Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, Kay NE, Brown JR, Gribben JG, Neuberg DS. et al.Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112(5):1923–1930. doi: 10.1182/blood-2007-05-092882. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

