A mathematical framework for analyzing T cell receptor scanning of peptides
- PMID: 21044568
- PMCID: PMC2966000
- DOI: 10.1016/j.bpj.2010.08.024
A mathematical framework for analyzing T cell receptor scanning of peptides
Abstract
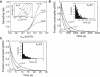
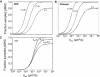
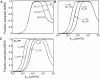
T cells continuously search for antigenic peptides presented on major histocompatibility complexes expressed on nearly all nucleated cells. Because only a few antigenic peptides are presented in a sea of thousands of self-peptides, the T cells have a critical task in discriminating between self- and nonself-peptides. This search process for antigens must be performed with sufficient speed in order to induce a fast response against invading pathogens. This study presents a mathematical framework for analyzing the scanning process of peptides. The framework includes analytic expressions for calculating the sampling rate as well as continuous-systems- and stochastic-agent-based models. The results show that the scanning of self-peptides is a very fast process due to fast off-rates. The simulations also predict the existence of an optimal sampling rate for a certain range of on-rates based on the recently proposed confinement time model. Calculations reveal that most of the self-peptides located within a microdomain are scanned within just a few seconds, and that the T cell receptors have kinetics for self-peptides, facilitating fast scanning. The derived mathematical expressions within this study provide conceptual calculations for further investigations of how the T cell discriminates between self- and nonself-peptides.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
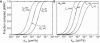

References
-
- Mahajan V.S., Leskov I.B., Chen J.Z. Homeostasis of T cell diversity. Cell. Mol. Immunol. 2005;2:1–10. - PubMed
-
- Alam S.M., Travers P.J., Gascoigne N.R. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. - PubMed
-
- Stoll S., Delon J., Germain R.N. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. - PubMed
-
- Irvine D.J., Purbhoo M.A., Davis M.M. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

