Single-molecule study of ribosome hierarchic dynamics at the peptidyl transferase center
- PMID: 21044598
- PMCID: PMC2966008
- DOI: 10.1016/j.bpj.2010.08.037
Single-molecule study of ribosome hierarchic dynamics at the peptidyl transferase center
Abstract
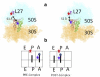
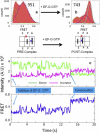
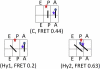
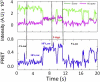
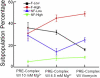
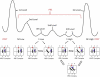
During protein biosynthesis the ribosome moves along mRNA in steps of precisely three nucleotides. The mechanism for this ribosome motion remains elusive. Using a classification algorithm to sort single-molecule fluorescence resonance energy transfer data into subpopulations, we found that the ribosome dynamics detected at the peptidyl transferase center are highly inhomogeneous. The pretranslocation complex has at least four subpopulations that sample two hybrid states, whereas the posttranslocation complex is mainly static. We observed transitions among the ribosome subpopulations under various conditions, including 1), in the presence of EF-G; 2), spontaneously; 3), in different buffers, and 4), bound to antibiotics. Therefore, these subpopulations represent biologically active ribosomes. One key observation indicates that the Hy2 hybrid state only exists in a fluctuating ribosome subpopulation, which prompts us to propose that ribosome dynamics are hierarchically arranged. This proposal may have important implications for the regulation of cellular translation rates.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Schmeing T.M., Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. - PubMed
-
- Nierhaus K.H. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. - PubMed
-
- Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. - PubMed
-
- Bretscher M.S. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

