ThYme: a database for thioester-active enzymes
- PMID: 21045059
- PMCID: PMC3013676
- DOI: 10.1093/nar/gkq1072
ThYme: a database for thioester-active enzymes
Abstract
The ThYme (Thioester-active enzYme; http://www.enzyme.cbirc.iastate.edu) database has been constructed to bring together amino acid sequences and 3D (tertiary) structures of all the enzymes constituting the fatty acid synthesis and polyketide synthesis cycles. These enzymes are active on thioester-containing substrates, specifically those that are parts of the acyl-CoA synthase, acyl-CoA carboxylase, acyl transferase, ketoacyl synthase, ketoacyl reductase, hydroxyacyl dehydratase, enoyl reductase and thioesterase enzyme groups. These groups have been classified into families, members of which are similar in sequences, tertiary structures and catalytic mechanisms, implying common protein ancestry. ThYme is continually updated as sequences and tertiary structures become available.
Figures
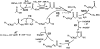

References
-
- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) Enzyme Nomenclature. Recommendations 1992. Academic Press, San Diego. http://www.chem.qmul.ac.uk/iubmb/
-
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

