Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code
- PMID: 21045127
- PMCID: PMC2993388
- DOI: 10.1073/pnas.1009023107
Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code
Abstract
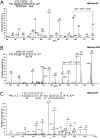
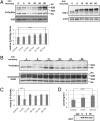
Dynamic posttranslational modification of serine and threonine residues of nucleocytoplasmic proteins by β-N-acetylglucosamine (O-GlcNAc) is a regulator of cellular processes such as transcription, signaling, and protein-protein interactions. Like phosphorylation, O-GlcNAc cycles in response to a wide variety of stimuli. Although cycling of O-GlcNAc is catalyzed by only two highly conserved enzymes, O-GlcNAc transferase (OGT), which adds the sugar, and β-N-acetylglucosaminidase (O-GlcNAcase), which hydrolyzes it, the targeting of these enzymes is highly specific and is controlled by myriad interacting subunits. Here, we demonstrate by multiple specific immunological and enzymatic approaches that histones, the proteins that package DNA within the nucleus, are O-GlcNAcylated in vivo. Histones also are substrates for OGT in vitro. We identify O-GlcNAc sites on histones H2A, H2B, and H4 using mass spectrometry. Finally, we show that histone O-GlcNAcylation changes during mitosis and with heat shock. Taken together, these data show that O-GlcNAc cycles dynamically on histones and can be considered part of the histone code.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Epigenetics gets sweeter: O-GlcNAc joins the "histone code".Chem Biol. 2010 Dec 22;17(12):1272-4. doi: 10.1016/j.chembiol.2010.12.001. Chem Biol. 2010. PMID: 21168762 Free PMC article.
References
-
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. - PubMed
-
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
-
- Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer's disease. Mol Biosyst. 2007;3:766–772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

