Roles of DEAD-box proteins in RNA and RNP Folding
- PMID: 21045543
- PMCID: PMC3073326
- DOI: 10.4161/rna.7.6.13571
Roles of DEAD-box proteins in RNA and RNP Folding
Abstract
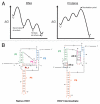
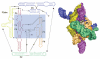
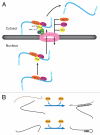
RNAs and RNA-protein complexes (RNPs) traverse rugged energy landscapes as they fold to their native structures, and many continue to undergo conformational rearrangements as they function. Due to the inherent stability of local RNA structure, proteins are required to assist with RNA conformational transitions during initial folding and in exchange between functional structures. DEAD-box proteins are superfamily 2 RNA helicases that are ubiquitously involved in RNA-mediated processes. Some of these proteins use an ATP-dependent cycle of conformational changes to disrupt RNA structure nonprocessively, accelerating structural transitions of RNAs and RNPs in a manner that bears a strong resemblance to the activities of certain groups of protein chaperones. This review summarizes recent work using model substrates and tractable self-splicing intron RNAs, which has given new insights into how DEAD-box proteins promote RNA folding steps and conformational transitions, and it summarizes recent progress in identifying sites and mechanisms of DEAD-box protein activity within more complex cellular targets.
Figures



Similar articles
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
-
Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs.Biochem Soc Trans. 2017 Dec 15;45(6):1313-1321. doi: 10.1042/BST20170095. Epub 2017 Nov 17. Biochem Soc Trans. 2017. PMID: 29150525 Free PMC article. Review.
-
DEAD-box proteins as RNA helicases and chaperones.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):135-52. doi: 10.1002/wrna.50. Wiley Interdiscip Rev RNA. 2011. PMID: 21297876 Free PMC article. Review.
-
Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones.J Mol Biol. 2009 Jun 19;389(4):674-93. doi: 10.1016/j.jmb.2009.04.043. Epub 2009 Apr 23. J Mol Biol. 2009. PMID: 19393667 Free PMC article.
-
Toward a molecular understanding of RNA remodeling by DEAD-box proteins.RNA Biol. 2013 Jan;10(1):44-55. doi: 10.4161/rna.22210. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995827 Free PMC article. Review.
Cited by
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
-
The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins.J Biol Chem. 2011 Oct 28;286(43):37304-12. doi: 10.1074/jbc.M111.287706. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878649 Free PMC article.
-
RNA chaperones buffer deleterious mutations in E. coli.Elife. 2015 Mar 25;4:e04745. doi: 10.7554/eLife.04745. Elife. 2015. PMID: 25806682 Free PMC article.
-
The DbpA catalytic core unwinds double-helix substrates by directly loading on them.RNA. 2016 Mar;22(3):408-15. doi: 10.1261/rna.052928.115. Epub 2016 Jan 11. RNA. 2016. PMID: 26755693 Free PMC article.
-
Analyzing ATP utilization by DEAD-Box RNA helicases using kinetic and equilibrium methods.Methods Enzymol. 2012;511:29-63. doi: 10.1016/B978-0-12-396546-2.00002-4. Methods Enzymol. 2012. PMID: 22713314 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
