Quantitative analysis of regulatory flexibility under changing environmental conditions
- PMID: 21045818
- PMCID: PMC3010117
- DOI: 10.1038/msb.2010.81
Quantitative analysis of regulatory flexibility under changing environmental conditions
Abstract
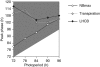
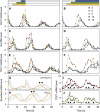
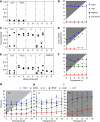
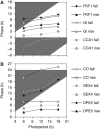
The circadian clock controls 24-h rhythms in many biological processes, allowing appropriate timing of biological rhythms relative to dawn and dusk. Known clock circuits include multiple, interlocked feedback loops. Theory suggested that multiple loops contribute the flexibility for molecular rhythms to track multiple phases of the external cycle. Clear dawn- and dusk-tracking rhythms illustrate the flexibility of timing in Ipomoea nil. Molecular clock components in Arabidopsis thaliana showed complex, photoperiod-dependent regulation, which was analysed by comparison with three contrasting models. A simple, quantitative measure, Dusk Sensitivity, was introduced to compare the behaviour of clock models with varying loop complexity. Evening-expressed clock genes showed photoperiod-dependent dusk sensitivity, as predicted by the three-loop model, whereas the one- and two-loop models tracked dawn and dusk, respectively. Output genes for starch degradation achieved dusk-tracking expression through light regulation, rather than a dusk-tracking rhythm. Model analysis predicted which biochemical processes could be manipulated to extend dusk tracking. Our results reveal how an operating principle of biological regulators applies specifically to the plant circadian clock.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Photoperiod-dependent changes in the phase of core clock transcripts and global transcriptional outputs at dawn and dusk in Arabidopsis.Plant Cell Environ. 2016 Sep;39(9):1955-81. doi: 10.1111/pce.12754. Epub 2016 Jul 15. Plant Cell Environ. 2016. PMID: 27075884
-
The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops.Mol Syst Biol. 2012 Mar 6;8:574. doi: 10.1038/msb.2012.6. Mol Syst Biol. 2012. PMID: 22395476 Free PMC article.
-
Light and circadian regulation of clock components aids flexible responses to environmental signals.New Phytol. 2014 Jul;203(2):568-577. doi: 10.1111/nph.12853. Epub 2014 May 20. New Phytol. 2014. PMID: 24842166 Free PMC article.
-
From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana.Biosci Biotechnol Biochem. 2013;77(1):10-6. doi: 10.1271/bbb.120765. Epub 2013 Jan 7. Biosci Biotechnol Biochem. 2013. PMID: 23291766 Review.
-
Circadian Rhythms in Plants.Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a034611. doi: 10.1101/cshperspect.a034611. Cold Spring Harb Perspect Biol. 2019. PMID: 31138544 Free PMC article. Review.
Cited by
-
The cyanobacterial circadian clock follows midday in vivo and in vitro.Elife. 2017 Jul 7;6:e23539. doi: 10.7554/eLife.23539. Elife. 2017. PMID: 28686160 Free PMC article.
-
The sugar-responsive circadian clock regulator bZIP63 modulates plant growth.New Phytol. 2021 Sep;231(5):1875-1889. doi: 10.1111/nph.17518. Epub 2021 Jun 30. New Phytol. 2021. PMID: 34053087 Free PMC article.
-
Circadian entrainment in Arabidopsis.Plant Physiol. 2022 Sep 28;190(2):981-993. doi: 10.1093/plphys/kiac204. Plant Physiol. 2022. PMID: 35512209 Free PMC article.
-
P19 Cells as a Model for Studying the Circadian Clock in Stem Cells before and after Cell Differentiation.J Circadian Rhythms. 2018 May 18;16:6. doi: 10.5334/jcr.157. J Circadian Rhythms. 2018. PMID: 30210566 Free PMC article.
-
Study on differentially expressed genes related to defoliation traits in two alfalfa varieties based on RNA-Seq.BMC Genomics. 2018 Nov 7;19(1):807. doi: 10.1186/s12864-018-5180-1. BMC Genomics. 2018. PMID: 30404602 Free PMC article.
References
-
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 - PubMed
-
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D019621/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- E015263/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G19886/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E015263/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G19886/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

