Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide
- PMID: 21045820
- PMCID: PMC3010120
- DOI: 10.1038/msb.2010.79
Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide
Abstract
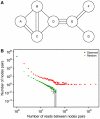
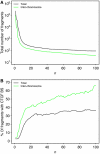
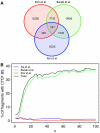
A prime goal in systems biology is the comprehensive use of existing high-throughput genomic datasets to gain a better understanding of chromatin organization and genome function. In this report, we use chromatin immunoprecipitation (ChIP) data that map protein-binding sites on the genome, and Hi-C data that map interactions between DNA fragments in the genome in an integrative approach. We first reanalyzed the contact map of the human genome as determined with Hi-C and found that long-range interactions are highly nonrandom; the same DNA fragments are often found interacting together. We then show using ChIP data that these interactions can be explained by the action of the CCCTC-binding factor (CTCF). These CTCF-mediated interactions are found both within chromosomes and in between different chromosomes. This makes CTCF a major organizer of both the structure of the chromosomal fiber within each individual chromosome and of the chromosome territories within the cell nucleus.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- de Laat W, Grosveld F (2007) Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev 17: 456–464 - PubMed
-
- Hertz GZ, Stormo GD (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563–577 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

