The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association
- PMID: 21047799
- PMCID: PMC3061077
- DOI: 10.1093/nar/gkq900
The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association
Abstract
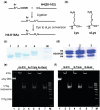
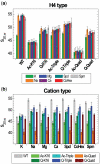
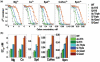
Understanding the molecular mechanisms behind regulation of chromatin folding through covalent modifications of the histone N-terminal tails is hampered by a lack of accessible chromatin containing precisely modified histones. We study the internal folding and intermolecular self-association of a chromatin system consisting of saturated 12-mer nucleosome arrays containing various combinations of completely acetylated lysines at positions 5, 8, 12 and 16 of histone H4, induced by the cations Na(+), K(+), Mg(2+), Ca(2+), cobalt-hexammine(3+), spermidine(3+) and spermine(4+). Histones were prepared using a novel semi-synthetic approach with native chemical ligation. Acetylation of H4-K16, but not its glutamine mutation, drastically reduces cation-induced folding of the array. Neither acetylations nor mutations of all the sites K5, K8 and K12 can induce a similar degree of array unfolding. The ubiquitous K(+), (as well as Rb(+) and Cs(+)) showed an unfolding effect on unmodified arrays almost similar to that of H4-K16 acetylation. We propose that K(+) (and Rb(+)/Cs(+)) binding to a site on the H2B histone (R96-L99) disrupts H4K16 ε-amino group binding to this specific site, thereby deranging H4 tail-mediated nucleosome-nucleosome stacking and that a similar mechanism operates in the case of H4-K16 acetylation. Inter-array self-association follows electrostatic behavior and is largely insensitive to the position or nature of the H4 tail charge modification.
Figures




References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Cryst. Sect. D. 2000;56:1513–1534. - PubMed
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. - PubMed
-
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

