Systems biology and genomics of breast cancer
- PMID: 21047916
- PMCID: PMC3039533
- DOI: 10.1101/cshperspect.a003293
Systems biology and genomics of breast cancer
Abstract
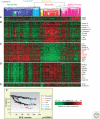
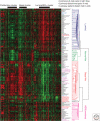
It is now accepted that breast cancer is not a single disease, but instead it is composed of a spectrum of tumor subtypes with distinct cellular origins, somatic changes, and etiologies. Gene expression profiling using DNA microarrays has contributed significantly to our understanding of the molecular heterogeneity of breast tumor formation, progression, and recurrence. For example, at least two clinical diagnostic assays exist (i.e., OncotypeDX RS and Mammaprint®) that are able to predict outcome in patients using patterns of gene expression and predetermined mathematical algorithms. In addition, a new molecular taxonomy based upon the inherent, or "intrinsic," biology of breast tumors has been developed; this taxonomy is called the "intrinsic subtypes of breast cancer," which now identifies five distinct tumor types and a normal breast-like group. Importantly, the intrinsic subtypes of breast cancer predict patient relapse, overall survival, and response to endocrine and chemotherapy regimens. Thus, most of the clinical behavior of a breast tumor is already written in its subtype profile. Here, we describe the discovery and basic biology of the intrinsic subtypes of breast cancer, and detail how this interacts with underlying genetic alternations, response to therapy, and the metastatic process.
Figures
References
-
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO 2004. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 203: 661–671 - PubMed
-
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A, Medina D 2008. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res 14: 370–378 - PubMed
-
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451 - PubMed
-
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2006. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
