A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait
- PMID: 21047983
- PMCID: PMC3022402
- DOI: 10.1093/jxb/erq346
A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait
Abstract
Genes that enable crops to limit Na(+) accumulation in shoot tissues represent potential sources of salinity tolerance for breeding. In barley, the HvNax4 locus lowered shoot Na(+) content by between 12% and 59% (g(-1) DW), or not at all, depending on the growth conditions in hydroponics and a range of soil types, indicating a strong influence of environment on expression. HvNax4 was fine-mapped on the long arm of barley chromosome 1H. Corresponding intervals of ∼200 kb, containing a total of 34 predicted genes, were defined in the sequenced rice and Brachypodium genomes. HvCBL4, a close barley homologue of the SOS3 salinity tolerance gene of Arabidopsis, co-segregated with HvNax4. No difference in HvCBL4 mRNA expression was detected between the mapping parents. However, genomic and cDNA sequences of the HvCBL4 alleles were obtained, revealing a single Ala111Thr amino acid substitution difference in the encoded proteins. The known crystal structure of SOS3 was used as a template to obtain molecular models of the barley proteins, resulting in structures very similar to that of SOS3. The position in SOS3 corresponding to the barley substitution does not participate directly in Ca(2+) binding, post-translational modifications or interaction with the SOS2 signalling partner. However, Thr111 but not Ala111 forms a predicted hydrogen bond with a neighbouring α-helix, which has potential implications for the overall structure and function of the barley protein. HvCBL4 therefore represents a candidate for HvNax4 that warrants further investigation.
Figures






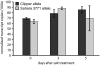
References
-
- Batistič O, Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochimica et Biophysica Acta. 2009;1793:985–992. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

