Re-examination of CD91 function in GRP94 (glycoprotein 96) surface binding, uptake, and peptide cross-presentation
- PMID: 21048103
- PMCID: PMC3717329
- DOI: 10.4049/jimmunol.1000448
Re-examination of CD91 function in GRP94 (glycoprotein 96) surface binding, uptake, and peptide cross-presentation
Abstract
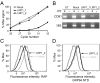
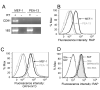
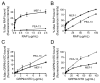
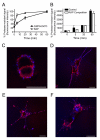
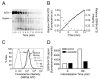
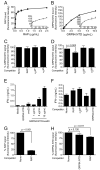
GRP94 (gp96)-peptide complexes can be internalized by APCs and their associated peptides cross-presented to yield activation of CD8(+) T cells. Investigations into the identity (or identities) of GRP94 surface receptors have yielded conflicting results, particularly with respect to CD91 (LRP1), which has been proposed to be essential for GRP94 recognition and uptake. To assess CD91 function in GRP94 surface binding and endocytosis, these parameters were examined in mouse embryonic fibroblast (MEF) cell lines whose expression of CD91 was either reduced via RNA interference or eliminated by genetic disruption of the CD91 locus. Reduction or loss of CD91 expression abrogated the binding and uptake of receptor-associated protein, an established CD91 ligand. Surface binding and uptake of an N-terminal domain of GRP94 (GRP94.NTD) was unaffected. GRP94.NTD surface binding was markedly suppressed after treatment of MEF cell lines with heparin, sodium chlorate, or heparinase II, demonstrating that heparin sulfate proteoglycans can function in GRP94.NTD surface binding. The role of CD91 in the cross-presentation of GRP94-associated peptides was examined in the DC2.4 dendritic cell line. In DC2.4 cells, which express CD91, GRP94.NTD-peptide cross-presentation was insensitive to the CD91 ligands receptor-associated protein or activated α(2)-macroglobulin and occurred primarily via a fluid-phase, rather than receptor-mediated, uptake pathway. These data clarify conflicting data on CD91 function in GRP94 surface binding, endocytosis, and peptide cross-presentation and identify a role for heparin sulfate proteoglycans in GRP94 surface binding.
Figures

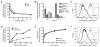
References
-
- Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185–194. - PubMed
-
- Nicchitta CV. Re-evaluating the role of heat-shock protein-peptide interactions in tumour immunity. Nat Rev Immunol. 2003;3:427–432. - PubMed
-
- Binder RJ, Harris ML, Menoret A, Srivastava PK. Saturation, competition, and specificity in interaction of heat shock proteins (hsp) gp96, hsp90, and hsp70 with CD11b+ cells. J Immunol. 2000;165:2582–2587. - PubMed
-
- Wassenberg JJ, Dezfulian C, Nicchitta CV. Receptor mediated and fluid phase pathways for internalization of the ER Hsp90 chaperone GRP94 in murine macrophages. J Cell Sci. 1999;112(Pt 13):2167–2175. - PubMed
-
- Arnold-Schild D, Hanau D, Spehner D, Schmid C, Rammensee HG, de la Salle H, Schild H. Cutting edge: receptor-mediated endocytosis of heat shock proteins by professional antigen-presenting cells. J Immunol. 1999;162:3757–3760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

