Mammalian circadian clock and metabolism - the epigenetic link
- PMID: 21048160
- PMCID: PMC2972271
- DOI: 10.1242/jcs.051649
Mammalian circadian clock and metabolism - the epigenetic link
Abstract
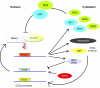
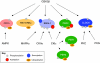
Circadian rhythms regulate a wide variety of physiological and metabolic processes. The clock machinery comprises complex transcriptional-translational feedback loops that, through the action of specific transcription factors, modulate the expression of as many as 10% of cellular transcripts. This marked change in gene expression necessarily implicates a global regulation of chromatin remodeling. Indeed, various descriptive studies have indicated that histone modifications occur at promoters of clock-controlled genes (CCGs) in a circadian manner. The finding that CLOCK, a transcription factor crucial for circadian function, has intrinsic histone acetyl transferase (HAT) activity has paved the way to unraveling the molecular mechanisms that govern circadian chromatin remodeling. A search for the histone deacetylase (HDAC) that counterbalances CLOCK activity revealed that SIRT1, a nicotinamide adenin dinucleotide (NAD(+))-dependent HDAC, functions in a circadian manner. Importantly, SIRT1 is a regulator of aging, inflammation and metabolism. As many transcripts that oscillate in mammalian peripheral tissues encode proteins that have central roles in metabolic processes, these findings establish a functional and molecular link between energy balance, chromatin remodeling and circadian physiology. Here we review recent studies that support the existence of this link and discuss their implications for understanding mammalian physiology and pathology.
Figures

Similar articles
-
The time of metabolism: NAD+, SIRT1, and the circadian clock.Cold Spring Harb Symp Quant Biol. 2011;76:31-8. doi: 10.1101/sqb.2011.76.010520. Epub 2011 Dec 16. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22179986 Review.
-
Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1.Int J Biochem Cell Biol. 2009 Jan;41(1):81-6. doi: 10.1016/j.biocel.2008.08.035. Epub 2008 Sep 4. Int J Biochem Cell Biol. 2009. PMID: 18817890 Review.
-
Minireview: NAD+, a circadian metabolite with an epigenetic twist.Endocrinology. 2012 Jan;153(1):1-5. doi: 10.1210/en.2011-1535. Epub 2011 Dec 20. Endocrinology. 2012. PMID: 22186411 Free PMC article. Review.
-
The epigenetic language of circadian clocks.Handb Exp Pharmacol. 2013;(217):29-44. doi: 10.1007/978-3-642-25950-0_2. Handb Exp Pharmacol. 2013. PMID: 23604474 Review.
-
Coupling circadian rhythms of metabolism and chromatin remodelling.Diabetes Obes Metab. 2015 Sep;17 Suppl 1(0 1):17-22. doi: 10.1111/dom.12509. Diabetes Obes Metab. 2015. PMID: 26332964 Free PMC article. Review.
Cited by
-
Sirtuin 1-mediated cellular metabolic memory of high glucose via the LKB1/AMPK/ROS pathway and therapeutic effects of metformin.Diabetes. 2012 Jan;61(1):217-28. doi: 10.2337/db11-0416. Epub 2011 Nov 28. Diabetes. 2012. PMID: 22124463 Free PMC article.
-
Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation.Cell Rep. 2018 Nov 13;25(7):1816-1828.e4. doi: 10.1016/j.celrep.2018.10.068. Cell Rep. 2018. PMID: 30428350 Free PMC article.
-
Long-Term Effects of Altered Photoperiod During Pregnancy on Liver Gene Expression of the Progeny.Front Physiol. 2019 Nov 22;10:1377. doi: 10.3389/fphys.2019.01377. eCollection 2019. Front Physiol. 2019. PMID: 31824324 Free PMC article.
-
The Emerging Role of Chromatin Remodeling Factors in Female Pubertal Development.Neuroendocrinology. 2019;109(3):208-217. doi: 10.1159/000497745. Epub 2019 Feb 7. Neuroendocrinology. 2019. PMID: 30731454 Free PMC article. Review.
-
Thyroxine differentially modulates the peripheral clock: lessons from the human hair follicle.PLoS One. 2015 Mar 30;10(3):e0121878. doi: 10.1371/journal.pone.0121878. eCollection 2015. PLoS One. 2015. PMID: 25822259 Free PMC article.
References
-
- Akashi M., Takumi T. (2005). The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12, 441-448 - PubMed
-
- Akhtar R. A., Reddy A. B., Maywood E. S., Clayton J. D., King V. M., Smith A. G., Gant T. W., Hastings M. H., Kyriacou C. P. (2002). Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540-550 - PubMed
-
- Akten B., Jauch E., Genova G. K., Kim E. Y., Edery I., Raabe T., Jackson F. R. (2003). A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6, 251-257 - PubMed
-
- Ando H., Yanagihara H., Hayashi Y., Obi Y., Tsuruoka S., Takamura T., Kaneko S., Fujimura A. (2005). Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology 146, 5631-5636 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

