Cytochrome P450 102A2 Catalyzes Efficient Oxidation of Sodium Dodecyl Sulphate: A Molecular Tool for Remediation
- PMID: 21048857
- PMCID: PMC2956967
- DOI: 10.4061/2010/125429
Cytochrome P450 102A2 Catalyzes Efficient Oxidation of Sodium Dodecyl Sulphate: A Molecular Tool for Remediation
Abstract
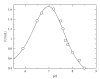
Bacterial cytochrome P450s (CYPs) constitute an important family of monooxygenase enzymes that carry out essential roles in the metabolism of endogenous compounds and foreign chemicals. In the present work we report the characterization of CYP102A2 from B. subtilis with a focus on its substrate specificity. CYP102A2 is more active in oxidation of sodium dodecyl sulphate (SDS) than any other characterized CYP. The effect of SDS and NADPH concentration on reaction rate showed nonhyperbolic and hyperbolic dependence, respectively. The enzyme was found to exhibit a bell-shaped curve for plots of activity versus pH, over pH values 5.9-8.5. The rate of SDS oxidation reached the maximum value approximately at pH 7.2 and the pH transition observed controlled by two pK(a)s in the acidic (pK(a) = 6.7 ± 0.08) and basic (pK(a) = 7.3 ± 0.06) pH range. The results are discussed in relation to the future biotechnology applications of CYPs.
Figures




References
-
- Narhi LO, Fulco AJ. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium . The Journal of Biological Chemistry. 1986;261(16):7160–7169. - PubMed
-
- Nelson DR, Koymans L, Kamataki T, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6(1):1–42. - PubMed
-
- Li H, Poulos TL. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nature Structural Biology. 1997;4(2):140–146. - PubMed
-
- Wen LP, Fulco AJ. Cloning of the gene encoding a catalytically self-sufficient cytochrome P-450 fatty acid monooxygenase induced by barbiturates in Bacillus megaterium and its functional expression and regulation in heterologous (Escherichia coli) and homologous (Bacillus megaterium) hosts. The Journal of Biological Chemistry. 1987;262(14):6676–6682. - PubMed
-
- Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson JA, Deisenhofer J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450’s. Science. 1993;261(5122):731–736. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

