Kinetics of Methylation by EcoP1I DNA Methyltransferase
- PMID: 21048863
- PMCID: PMC2962900
- DOI: 10.4061/2010/302731
Kinetics of Methylation by EcoP1I DNA Methyltransferase
Abstract
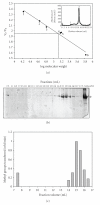
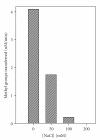
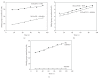
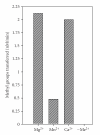
EcoP1I DNA MTase (M.EcoP1I), an N(6)-adenine MTase from bacteriophage P1, is a part of the EcoP1I restriction-modification (R-M) system which belongs to the Type III R-M system. It recognizes the sequence 5'-AGACC-3' and methylates the internal adenine. M.EcoP1I requires Mg(2+) for the transfer of methyl groups to DNA. M.EcoP1I is shown to exist as dimer in solution, and even at high salt concentrations (0.5 M) the dimeric M.EcoP1I does not dissociate into monomers suggesting a strong interaction between the monomer subunits. Preincubation and isotope partitioning studies with M.EcoP1I indicate a kinetic mechanism where the duplex DNA binds first followed by AdoMet. Interestingly, M.EcoP1I methylates DNA substrates in the presence of Mn(2+) and Ca(2+) other than Mg(2+) with varying affinities. Amino acid analysis and methylation assays in the presence of metal ions suggest that M.EcoP1I has indeed two metal ion-binding sites [(358)ID(x)(n) … ExK(401) and (600)DxDxD(604) motif]. EcoP1I DNA MTase catalyzes the transfer of methyl groups using a distributive mode of methylation on DNA containing more than one recognition site. A chemical modification of EcoP1I DNA MTase using N-ethylmaleimide resulted in an irreversible inactivation of enzyme activity suggesting the possible role of cysteine residues in catalysis.
Figures



References
-
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. Journal of Molecular Biology. 1995;253(4):618–632. - PubMed
-
- Hadi SM, Bächi B, Iida S, Bickle TA. DNA restriction-modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. Journal of Molecular Biology. 1983;165(1):19–34. - PubMed
-
- Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84(11):1047–1059. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

