Genome wide association studies for milk production traits in Chinese Holstein population
- PMID: 21048968
- PMCID: PMC2965099
- DOI: 10.1371/journal.pone.0013661
Genome wide association studies for milk production traits in Chinese Holstein population
Abstract
Genome-wide association studies (GWAS) based on high throughput SNP genotyping technologies open a broad avenue for exploring genes associated with milk production traits in dairy cattle. Motivated by pinpointing novel quantitative trait nucleotide (QTN) across Bos Taurus genome, the present study is to perform GWAS to identify genes affecting milk production traits using current state-of-the-art SNP genotyping technology, i.e., the Illumina BovineSNP50 BeadChip. In the analyses, the five most commonly evaluated milk production traits are involved, including milk yield (MY), milk fat yield (FY), milk protein yield (PY), milk fat percentage (FP) and milk protein percentage (PP). Estimated breeding values (EBVs) of 2,093 daughters from 14 paternal half-sib families are considered as phenotypes within the framework of a daughter design. Association tests between each trait and the 54K SNPs are achieved via two different analysis approaches, a paternal transmission disequilibrium test (TDT)-based approach (L1-TDT) and a mixed model based regression analysis (MMRA). In total, 105 SNPs were detected to be significantly associated genome-wise with one or multiple milk production traits. Of the 105 SNPs, 38 were commonly detected by both methods, while four and 63 were solely detected by L1-TDT and MMRA, respectively. The majority (86 out of 105) of the significant SNPs is located within the reported QTL regions and some are within or close to the reported candidate genes. In particular, two SNPs, ARS-BFGL-NGS-4939 and BFGL-NGS-118998, are located close to the DGAT1 gene (160bp apart) and within the GHR gene, respectively. Our findings herein not only provide confirmatory evidences for previously findings, but also explore a suite of novel SNPs associated with milk production traits, and thus form a solid basis for eventually unraveling the causal mutations for milk production traits in dairy cattle.
Conflict of interest statement
Figures
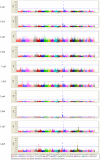
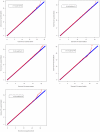
Similar articles
-
Genomic Regions and Candidate Genes Associated with Milk Production Traits in Holstein and Its Crossbred Cattle: A Review.Int J Genomics. 2023 Jul 27;2023:8497453. doi: 10.1155/2023/8497453. eCollection 2023. Int J Genomics. 2023. PMID: 37547753 Free PMC article. Review.
-
Short communication: Replication of genome-wide association studies for milk production traits in Chinese Holstein by an efficient rotated linear mixed model.J Dairy Sci. 2019 Mar;102(3):2378-2383. doi: 10.3168/jds.2018-15298. Epub 2019 Jan 11. J Dairy Sci. 2019. PMID: 30639022
-
A candidate gene association study for nine economically important traits in Italian Holstein cattle.Anim Genet. 2014 Aug;45(4):576-80. doi: 10.1111/age.12164. Epub 2014 May 2. Anim Genet. 2014. PMID: 24796806
-
Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein.PLoS One. 2014 May 23;9(5):e96186. doi: 10.1371/journal.pone.0096186. eCollection 2014. PLoS One. 2014. PMID: 24858810 Free PMC article.
-
Genetic mechanisms underlying spermatic and testicular traits within and among cattle breeds: systematic review and prioritization of GWAS results.J Anim Sci. 2018 Dec 3;96(12):4978-4999. doi: 10.1093/jas/sky382. J Anim Sci. 2018. PMID: 30304443 Free PMC article.
Cited by
-
Genome-wide association studies for growth and meat production traits in sheep.PLoS One. 2013 Jun 25;8(6):e66569. doi: 10.1371/journal.pone.0066569. Print 2013. PLoS One. 2013. PMID: 23825544 Free PMC article.
-
Genome-wide association for milk production and female fertility traits in Canadian dairy Holstein cattle.BMC Genet. 2016 Jun 10;17(1):75. doi: 10.1186/s12863-016-0386-1. BMC Genet. 2016. PMID: 27287773 Free PMC article.
-
Detection of selection signatures in dairy and beef cattle using high-density genomic information.Genet Sel Evol. 2015 Jun 19;47(1):49. doi: 10.1186/s12711-015-0127-3. Genet Sel Evol. 2015. PMID: 26089079 Free PMC article.
-
Genome-Wide Association Study for Lactation Performance in the Early and Peak Stages of Lactation in Holstein Dairy Cows.Animals (Basel). 2022 Jun 14;12(12):1541. doi: 10.3390/ani12121541. Animals (Basel). 2022. PMID: 35739877 Free PMC article.
-
Genomic Regions and Candidate Genes Associated with Milk Production Traits in Holstein and Its Crossbred Cattle: A Review.Int J Genomics. 2023 Jul 27;2023:8497453. doi: 10.1155/2023/8497453. eCollection 2023. Int J Genomics. 2023. PMID: 37547753 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

