Specified species in gingival crevicular fluid predict bacterial diversity
- PMID: 21049043
- PMCID: PMC2963608
- DOI: 10.1371/journal.pone.0013589
Specified species in gingival crevicular fluid predict bacterial diversity
Abstract
Background: Analysis of gingival crevicular fluid (GCF) samples may give information of unattached (planktonic) subgingival bacteria. Our study represents the first one targeting the identity of bacteria in GCF.
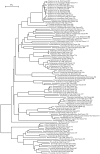
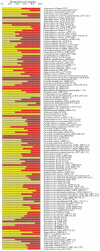
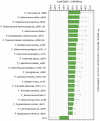
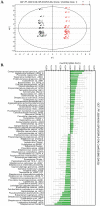
Methodology/principal findings: We determined bacterial species diversity in GCF samples of a group of periodontitis patients and delineated contributing bacterial and host-associated factors. Subgingival paper point (PP) samples from the same sites were taken for comparison. After DNA extraction, 16S rRNA genes were PCR amplified and DNA-DNA hybridization was performed using a microarray for over 300 bacterial species or groups. Altogether 133 species from 41 genera and 8 phyla were detected with 9 to 62 and 18 to 64 species in GCF and PP samples, respectively, per patient. Projection to latent structures by means of partial least squares (PLS) was applied to the multivariate data analysis. PLS regression analysis showed that species of genera including Campylobacter, Selenomonas, Porphyromonas, Catonella, Tannerella, Dialister, Peptostreptococcus, Streptococcus and Eubacterium had significant positive correlations and the number of teeth with low-grade attachment loss a significant negative correlation to species diversity in GCF samples. OPLS/O2PLS discriminant analysis revealed significant positive correlations to GCF sample group membership for species of genera Campylobacter, Leptotrichia, Prevotella, Dialister, Tannerella, Haemophilus, Fusobacterium, Eubacterium, and Actinomyces.
Conclusions/significance: Among a variety of detected species those traditionally classified as Gram-negative anaerobes growing in mature subgingival biofilms were the main predictors for species diversity in GCF samples as well as responsible for distinguishing GCF samples from PP samples. GCF bacteria may provide new prospects for studying dynamic properties of subgingival biofilms.
Conflict of interest statement
Figures


Similar articles
-
Association of early onset periodontitis microbiota with aspartate aminotransferase activity in gingival crevicular fluid.J Clin Periodontol. 2001 Dec;28(12):1096-105. doi: 10.1034/j.1600-051x.2001.281203.x. J Clin Periodontol. 2001. PMID: 11737506
-
Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray.J Periodontol. 2009 Sep;80(9):1421-32. doi: 10.1902/jop.2009.090185. J Periodontol. 2009. PMID: 19722792 Free PMC article.
-
Impact of aging on TREM-1 responses in the periodontium: a cross-sectional study in an elderly population.BMC Infect Dis. 2016 Aug 19;16(1):429. doi: 10.1186/s12879-016-1778-6. BMC Infect Dis. 2016. PMID: 27542376 Free PMC article.
-
Relationship of the subgingival microbiota to a chairside test for aspartate aminotransferase in gingival crevicular fluid.J Periodontol. 1999 Jan;70(1):57-62. doi: 10.1902/jop.1999.70.1.57. J Periodontol. 1999. PMID: 10052771
-
Comparative analysis of bacterial abundance and diversity in tumour tissue of oral squamous cell carcinoma and non-tumour tissue: insights from a systematic review of 16S ribosomal RNA sequencing.BMC Oral Health. 2025 Apr 16;25(1):577. doi: 10.1186/s12903-025-05941-3. BMC Oral Health. 2025. PMID: 40241078 Free PMC article.
Cited by
-
Impact of spacers and thermocycling on porosity and gaps in class II endodontic temporary restorations evaluated by microcomputed tomography.Sci Rep. 2025 Jun 6;15(1):19874. doi: 10.1038/s41598-025-03458-x. Sci Rep. 2025. PMID: 40481043 Free PMC article.
-
Lessons learned and unlearned in periodontal microbiology.Periodontol 2000. 2013 Jun;62(1):95-162. doi: 10.1111/prd.12010. Periodontol 2000. 2013. PMID: 23574465 Free PMC article. Review.
-
Leptotrichia species in human infections II.J Oral Microbiol. 2017 Sep 15;9(1):1368848. doi: 10.1080/20002297.2017.1368848. eCollection 2017. J Oral Microbiol. 2017. PMID: 29081911 Free PMC article. Review.
-
Metagenomic biomarker discovery and explanation.Genome Biol. 2011 Jun 24;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. Genome Biol. 2011. PMID: 21702898 Free PMC article.
-
Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay.J Clin Med. 2020 Sep 11;9(9):2945. doi: 10.3390/jcm9092945. J Clin Med. 2020. PMID: 32933084 Free PMC article.
References
-
- Baelum V, van Palenstein Helderman W, Hugoson A, Yee R, Fejerskov O. A global perspective on changes in the burden of caries and periodontitis: implications for dentistry. J Oral Rehabil. 2007;34:872–906; discussion 940. - PubMed
-
- Loos BG. Systemic markers of inflammation in periodontitis. J Periodontol. 2005;76:2106–2115. - PubMed
-
- Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. - PubMed
-
- Ridker PM, Silvertown JD. Inflammation, C-reactive protein, and atherothrombosis. J Periodontol. 2008;79:1544–1551. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

