The threshold bootstrap clustering: a new approach to find families or transmission clusters within molecular quasispecies
- PMID: 21049051
- PMCID: PMC2963616
- DOI: 10.1371/journal.pone.0013619
The threshold bootstrap clustering: a new approach to find families or transmission clusters within molecular quasispecies
Abstract
Background: Phylogenetic methods produce hierarchies of molecular species, inferring knowledge about taxonomy and evolution. However, there is not yet a consensus methodology that provides a crisp partition of taxa, desirable when considering the problem of intra/inter-patient quasispecies classification or infection transmission event identification. We introduce the threshold bootstrap clustering (TBC), a new methodology for partitioning molecular sequences, that does not require a phylogenetic tree estimation.
Methodology/principal findings: The TBC is an incremental partition algorithm, inspired by the stochastic Chinese restaurant process, and takes advantage of resampling techniques and models of sequence evolution. TBC uses as input a multiple alignment of molecular sequences and its output is a crisp partition of the taxa into an automatically determined number of clusters. By varying initial conditions, the algorithm can produce different partitions. We describe a procedure that selects a prime partition among a set of candidate ones and calculates a measure of cluster reliability. TBC was successfully tested for the identification of type-1 human immunodeficiency and hepatitis C virus subtypes, and compared with previously established methodologies. It was also evaluated in the problem of HIV-1 intra-patient quasispecies clustering, and for transmission cluster identification, using a set of sequences from patients with known transmission event histories.
Conclusion: TBC has been shown to be effective for the subtyping of HIV and HCV, and for identifying intra-patient quasispecies. To some extent, the algorithm was able also to infer clusters corresponding to events of infection transmission. The computational complexity of TBC is quadratic in the number of taxa, lower than other established methods; in addition, TBC has been enhanced with a measure of cluster reliability. The TBC can be useful to characterise molecular quasipecies in a broad context.
Conflict of interest statement
Figures

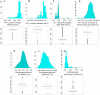

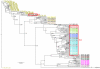
References
-
- Lemey P, Salemi M, Vandamme A-M, editors. NY: Cambridge University Press; 2009. The Phylogenetic Handbook: A Practical Approach to phylogenetic analysis and hypothesis testing.
-
- Felsenstein J. Sinauer Associates, Sunderland, MA; 2004. Inferring Phylogenies.
-
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–84. - PubMed
-
- Hendy MD, Penny D. Branch and bound algorithms to determine minimal evolutionary trees. Math Biosci. 1982;60:133–42.
-
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

