C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response
- PMID: 21049065
- PMCID: PMC2964293
- DOI: 10.1371/journal.pntd.0000856
C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response
Abstract
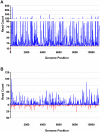
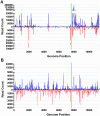
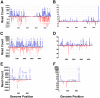
Mosquitoes rely on RNA interference (RNAi) as their primary defense against viral infections. To this end, the combination of RNAi and invertebrate cell culture systems has become an invaluable tool in studying virus-vector interactions. Nevertheless, a recent study failed to detect an active RNAi response to West Nile virus (WNV) infection in C6/36 (Aedes albopictus) cells, a mosquito cell line frequently used to study arthropod-borne viruses (arboviruses). Therefore, we sought to determine if WNV actively evades the host's RNAi response or if C6/36 cells have a dysfunctional RNAi pathway. C6/36 and Drosophila melanogaster S2 cells were infected with WNV (Flaviviridae), Sindbis virus (SINV, Togaviridae) and La Crosse virus (LACV, Bunyaviridae) and total RNA recovered from cell lysates. Small RNA (sRNA) libraries were constructed and subjected to high-throughput sequencing. In S2 cells, virus-derived small interfering RNAs (viRNAs) from all three viruses were predominantly 21 nt in length, a hallmark of the RNAi pathway. However, in C6/36 cells, viRNAs were primarily 17 nt in length from WNV infected cells and 26-27 nt in length in SINV and LACV infected cells. Furthermore, the origin (positive or negative viral strand) and distribution (position along viral genome) of S2 cell generated viRNA populations was consistent with previously published studies, but the profile of sRNAs isolated from C6/36 cells was altered. In total, these results suggest that C6/36 cells lack a functional antiviral RNAi response. These findings are analogous to the type-I interferon deficiency described in Vero (African green monkey kidney) cells and suggest that C6/36 cells may fail to accurately model mosquito-arbovirus interactions at the molecular level.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

