Quiescent fibroblasts exhibit high metabolic activity
- PMID: 21049082
- PMCID: PMC2958657
- DOI: 10.1371/journal.pbio.1000514
Quiescent fibroblasts exhibit high metabolic activity
Abstract
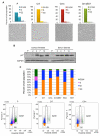
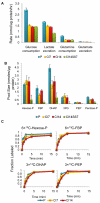
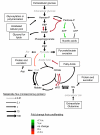
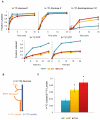
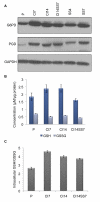
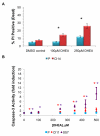
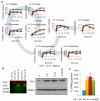
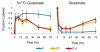
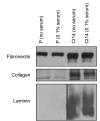
Many cells in mammals exist in the state of quiescence, which is characterized by reversible exit from the cell cycle. Quiescent cells are widely reported to exhibit reduced size, nucleotide synthesis, and metabolic activity. Much lower glycolytic rates have been reported in quiescent compared with proliferating lymphocytes. In contrast, we show here that primary human fibroblasts continue to exhibit high metabolic rates when induced into quiescence via contact inhibition. By monitoring isotope labeling through metabolic pathways and quantitatively identifying fluxes from the data, we show that contact-inhibited fibroblasts utilize glucose in all branches of central carbon metabolism at rates similar to those of proliferating cells, with greater overflow flux from the pentose phosphate pathway back to glycolysis. Inhibition of the pentose phosphate pathway resulted in apoptosis preferentially in quiescent fibroblasts. By feeding the cells labeled glutamine, we also detected a "backwards" flux in the tricarboxylic acid cycle from α-ketoglutarate to citrate that was enhanced in contact-inhibited fibroblasts; this flux likely contributes to shuttling of NADPH from the mitochondrion to cytosol for redox defense or fatty acid synthesis. The high metabolic activity of the fibroblasts was directed in part toward breakdown and resynthesis of protein and lipid, and in part toward excretion of extracellular matrix proteins. Thus, reduced metabolic activity is not a hallmark of the quiescent state. Quiescent fibroblasts, relieved of the biosynthetic requirements associated with generating progeny, direct their metabolic activity to preservation of self integrity and alternative functions beneficial to the organism as a whole.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
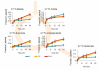
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

