Metabolomic analysis and visualization engine for LC-MS data
- PMID: 21049934
- PMCID: PMC5748896
- DOI: 10.1021/ac1021166
Metabolomic analysis and visualization engine for LC-MS data
Abstract
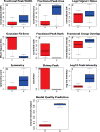
Metabolomic analysis by liquid chromatography-high-resolution mass spectrometry results in data sets with thousands of features arising from metabolites, fragments, isotopes, and adducts. Here we describe a software package, Metabolomic Analysis and Visualization ENgine (MAVEN), designed for efficient interactive analysis of LC-MS data, including in the presence of isotope labeling. The software contains tools for all aspects of the data analysis process, from feature extraction to pathway-based graphical data display. To facilitate data validation, a machine learning algorithm automatically assesses peak quality. Users interact with raw data primarily in the form of extracted ion chromatograms, which are displayed with overlaid circles indicating peak quality, and bar graphs of peak intensities for both unlabeled and isotope-labeled metabolite forms. Click-based navigation leads to additional information, such as raw data for specific isotopic forms or for metabolites changing significantly between conditions. Fast data processing algorithms result in nearly delay-free browsing. Drop-down menus provide tools for the overlay of data onto pathway maps. These tools enable animating series of pathway graphs, e.g., to show propagation of labeled forms through a metabolic network. MAVEN is released under an open source license at http://maven.princeton.edu.
Figures

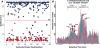



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

