Structure, energetics, and dynamics of binding coactivator peptide to the human retinoid X receptor α ligand binding domain complex with 9-cis-retinoic acid
- PMID: 21049972
- PMCID: PMC3081989
- DOI: 10.1021/bi101288y
Structure, energetics, and dynamics of binding coactivator peptide to the human retinoid X receptor α ligand binding domain complex with 9-cis-retinoic acid
Abstract
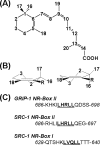
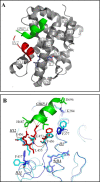
Retinoid X receptors (RXRs) are ligand-dependent nuclear receptors, which are activated by the potent agonist 9-cis-retinoic acid (9cRA). 9cRA binds to the ligand binding domain (LBD) of RXRs and recruits coactivator proteins for gene transcription. Using isothermal titration calorimetry, the binding of a 13-mer coactivator peptide, GRIP-1, to the hRXRα-LBD homodimer complex containing 9cRA (hRXRα-LBD:9cRA:GRIP-1) is reported between 20 and 37 °C. ΔG is temperature independent (-8.5 kcal/mol), and GRIP-1 binding is driven by ΔH (-9.2 kcal/mol) at 25 °C. ΔC(p) is large and negative (-401 cal mol(-1) K(-1)). The crystal structure of hRXRα-LBD:9cRA:GRIP-1 is reported at 2.05 Å. When the structures of hRXRα-LBD:9cRA:GRIP-1 and hRXRα-LBD:9cRA ( 1FBY ) homodimers are compared, E453 and E456 on helix 12 bury and form ionic interactions with GRIP-1. R302 on helix 4 realigns to form new salt bridges to both E453 and E456. F277 (helix 3), F437 (helix 11), and F450 (helix 12) move toward the hydrophobic interior. The changes in the near-UV spectrum at 260 nm of the hRXRα-LBD:9cRA:GRIP-1 support this structural change. Helix 11 tilts toward helix 12 by ≈1 Å, modifying the ring conformation of 9cRA. Hydrogen-deuterium exchange mass spectroscopy indicates GRIP-1 binding to hRXRα-LBD:9cRA significantly decreases the exchange rates for peptides containing helices 3 (F277), 4 (R302), 11 (F437), and 12 (E453, E456). The structural changes and loss of dynamics of the GRIP-1-bound structure are used to interpret the energetics of coactivator peptide binding to the agonist-bound hRXRα-LBD.
Figures





References
-
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. - PubMed
-
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. - PubMed
-
- Dong D, Noy N. Heterodimer formation by retinoid X receptor: regulation by ligands and by the receptor's self-association properties. Biochemistry. 1998;37:10691–10700. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

