Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules
- PMID: 21050467
- PMCID: PMC2991308
- DOI: 10.1186/1471-2407-10-604
Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules
Abstract
Background: Elucidating the activation pattern of molecular pathways across a given tumour type is a key challenge necessary for understanding the heterogeneity in clinical response and for developing novel more effective therapies. Gene expression signatures of molecular pathway activation derived from perturbation experiments in model systems as well as structural models of molecular interactions ("model signatures") constitute an important resource for estimating corresponding activation levels in tumours. However, relatively few strategies for estimating pathway activity from such model signatures exist and only few studies have used activation patterns of pathways to refine molecular classifications of cancer.
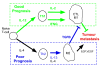
Methods: Here we propose a novel network-based method for estimating pathway activation in tumours from model signatures. We find that although the pathway networks inferred from cancer expression data are highly consistent with the prior information contained in the model signatures, that they also exhibit a highly modular structure and that estimation of pathway activity is dependent on this modular structure. We apply our methodology to a panel of 438 estrogen receptor negative (ER-) and 785 estrogen receptor positive (ER+) breast cancers to infer activation patterns of important cancer related molecular pathways.
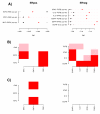
Results: We show that in ER negative basal and HER2+ breast cancer, gene expression modules reflecting T-cell helper-1 (Th1) and T-cell helper-2 (Th2) mediated immune responses play antagonistic roles as major risk factors for distant metastasis. Using Boolean interaction Cox-regression models to identify non-linear pathway combinations associated with clinical outcome, we show that simultaneous high activation of Th1 and low activation of a TGF-beta pathway module defines a subtype of particularly good prognosis and that this classification provides a better prognostic model than those based on the individual pathways. In ER+ breast cancer, we find that simultaneous high MYC and RAS activity confers significantly worse prognosis than either high MYC or high RAS activity alone. We further validate these novel prognostic classifications in independent sets of 173 ER- and 567 ER+ breast cancers.
Conclusion: We have proposed a novel method for pathway activity estimation in tumours and have shown that pathway modules antagonize or synergize to delineate novel prognostic subtypes. Specifically, our results suggest that simultaneous modulation of T-helper differentiation and TGF-beta pathways may improve clinical outcome of hormone insensitive breast cancers over treatments that target only one of these pathways.
Figures






Similar articles
-
An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer.Genome Biol. 2007;8(8):R157. doi: 10.1186/gb-2007-8-8-r157. Genome Biol. 2007. PMID: 17683518 Free PMC article.
-
Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures.Breast Cancer Res. 2008;10(4):R65. doi: 10.1186/bcr2124. Epub 2008 Jul 28. Breast Cancer Res. 2008. PMID: 18662380 Free PMC article.
-
Anti-HER2 CD4(+) T-helper type 1 response is a novel immune correlate to pathologic response following neoadjuvant therapy in HER2-positive breast cancer.Breast Cancer Res. 2015 May 23;17(1):71. doi: 10.1186/s13058-015-0584-1. Breast Cancer Res. 2015. PMID: 25997452 Free PMC article.
-
Molecular classification of estrogen receptor-positive/luminal breast cancers.Adv Anat Pathol. 2012 Jan;19(1):39-53. doi: 10.1097/PAP.0b013e31823fafa0. Adv Anat Pathol. 2012. PMID: 22156833 Review.
-
Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine.Am J Pathol. 2013 Oct;183(4):1113-1124. doi: 10.1016/j.ajpath.2013.08.002. Epub 2013 Aug 27. Am J Pathol. 2013. PMID: 23993780 Free PMC article. Review.
Cited by
-
MDR1-EXPRESSING CD4+ T CELLS WITH TH1.17 FEATURES RESIST TO NEOADJUVANT CHEMOTHERAPY AND ARE ASSOCIATED WITH BREAST CANCER CLINICAL RESPONSE.J Immunother Cancer. 2023 Nov;11(11):e007733. doi: 10.1136/jitc-2023-007733. J Immunother Cancer. 2023. PMID: 37940345 Free PMC article.
-
The immune contexture in human tumours: impact on clinical outcome.Nat Rev Cancer. 2012 Mar 15;12(4):298-306. doi: 10.1038/nrc3245. Nat Rev Cancer. 2012. PMID: 22419253 Review.
-
Gene expression and pathologic response to neoadjuvant chemotherapy in breast cancer.Mol Biol Rep. 2012 Jul;39(7):7435-41. doi: 10.1007/s11033-012-1576-1. Epub 2012 Feb 9. Mol Biol Rep. 2012. PMID: 22318550
-
Tumor-Infiltrating Lymphocytes (TILs) as a Biomarker of Abscopal Effect of Cryoablation in Breast Cancer: A Pilot Study.Ann Surg Oncol. 2022 May;29(5):2914-2925. doi: 10.1245/s10434-021-11157-w. Epub 2022 Jan 29. Ann Surg Oncol. 2022. PMID: 35094188 Free PMC article.
-
PD-L1 Expression and Tumor-infiltrating Lymphocytes in Breast Cancer: Clinicopathological Analysis in Women Younger than 40 Years Old.In Vivo. 2020 Mar-Apr;34(2):639-647. doi: 10.21873/invivo.11818. In Vivo. 2020. PMID: 32111764 Free PMC article.
References
-
- Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66:3903–3911. doi: 10.1158/0008-5472.CAN-05-4363. - DOI - PubMed
-
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR. mtor inhibition reverses akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and hif-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

