The endonuclease domain of MutL interacts with the β sliding clamp
- PMID: 21050827
- PMCID: PMC3028594
- DOI: 10.1016/j.dnarep.2010.10.003
The endonuclease domain of MutL interacts with the β sliding clamp
Abstract
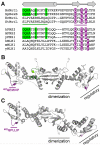
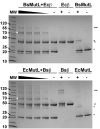
Mismatch repair corrects errors that have escaped polymerase proofreading enhancing replication fidelity by at least two orders of magnitude. The β and PCNA sliding clamps increase the polymerase processivity during DNA replication and are important at several stages of mismatch repair. Both MutS and MutL, the two proteins that initiate the mismatch repair response, interact with β. Binding of MutS to β is important to recruit MutS and MutL to foci. Moreover, the endonuclease activity of human and yeast MutLα is stimulated by PCNA. However, the concrete functions of the processivity clamp in the repair steps preceding DNA resynthesis remain obscure. Here, we demonstrate that the C-terminal domain of MutL encompasses a bona fide β-binding motif that mediates a weak, yet specific, interaction between the two proteins. Mutation of this conserved motif correlates with defects in mismatch repair, demonstrating that the direct interaction with β is important for MutL function. The interaction between the C-terminal domain of MutL and β is conserved in both Bacillus subtilis and Escherichia coli, but the repair defects associated with mutation of this β-binding motif are more severe in the former, suggesting that this interaction may have a more prominent role in methyl-independent than methyl-directed mismatch repair systems. Together with previously published data, our work strongly suggests that β may stimulate the endonuclease activity of MutL through its direct interaction with the C-terminal domain of MutL.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
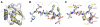


Similar articles
-
The sliding clamp tethers the endonuclease domain of MutL to DNA.Nucleic Acids Res. 2015 Dec 15;43(22):10746-59. doi: 10.1093/nar/gkv918. Epub 2015 Sep 17. Nucleic Acids Res. 2015. PMID: 26384423 Free PMC article.
-
Binding of the regulatory domain of MutL to the sliding β-clamp is species specific.Nucleic Acids Res. 2019 May 21;47(9):4831-4842. doi: 10.1093/nar/gkz115. Nucleic Acids Res. 2019. PMID: 30916336 Free PMC article.
-
The beta sliding clamp binds to multiple sites within MutL and MutS.J Biol Chem. 2006 May 19;281(20):14340-9. doi: 10.1074/jbc.M601264200. Epub 2006 Mar 16. J Biol Chem. 2006. PMID: 16546997
-
The functions of MutL in mismatch repair: the power of multitasking.Prog Mol Biol Transl Sci. 2012;110:41-70. doi: 10.1016/B978-0-12-387665-2.00003-1. Prog Mol Biol Transl Sci. 2012. PMID: 22749142 Review.
-
Insights from a decade of biophysical studies on MutL: Roles in strand discrimination and mismatch removal.Prog Biophys Mol Biol. 2015 Mar;117(2-3):149-156. doi: 10.1016/j.pbiomolbio.2015.02.002. Epub 2015 Feb 18. Prog Biophys Mol Biol. 2015. PMID: 25701376 Review.
Cited by
-
DNA repair and genome maintenance in Bacillus subtilis.Microbiol Mol Biol Rev. 2012 Sep;76(3):530-64. doi: 10.1128/MMBR.05020-11. Microbiol Mol Biol Rev. 2012. PMID: 22933559 Free PMC article. Review.
-
Yeast mutator phenotype enforced by Arabidopsis PMS1 expression.Mol Biol Rep. 2013 Mar;40(3):2107-14. doi: 10.1007/s11033-012-2269-5. Epub 2012 Nov 25. Mol Biol Rep. 2013. PMID: 23184005
-
DNA Mismatch Repair.EcoSal Plus. 2012 Nov;5(1):10.1128/ecosalplus.7.2.5. doi: 10.1128/ecosalplus.7.2.5. EcoSal Plus. 2012. PMID: 26442827 Free PMC article.
-
Experimental exchange of paralogous domains in the MLH family provides evidence of sub-functionalization after gene duplication.G3 (Bethesda). 2021 Jun 17;11(6):jkab111. doi: 10.1093/g3journal/jkab111. G3 (Bethesda). 2021. PMID: 33871573 Free PMC article.
-
pilE G-Quadruplex Is Recognized and Preferentially Bound but Not Processed by the MutL Endonuclease from Neisseria gonorrhoeae Mismatch Repair Pathway.Int J Mol Sci. 2023 Mar 24;24(7):6167. doi: 10.3390/ijms24076167. Int J Mol Sci. 2023. PMID: 37047138 Free PMC article.
References
-
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. - PubMed
-
- Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. - PubMed
-
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. - PubMed
-
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. - PubMed
-
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

