Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes
- PMID: 21050894
- PMCID: PMC3108001
- DOI: 10.1016/j.nano.2010.10.004
Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes
Abstract
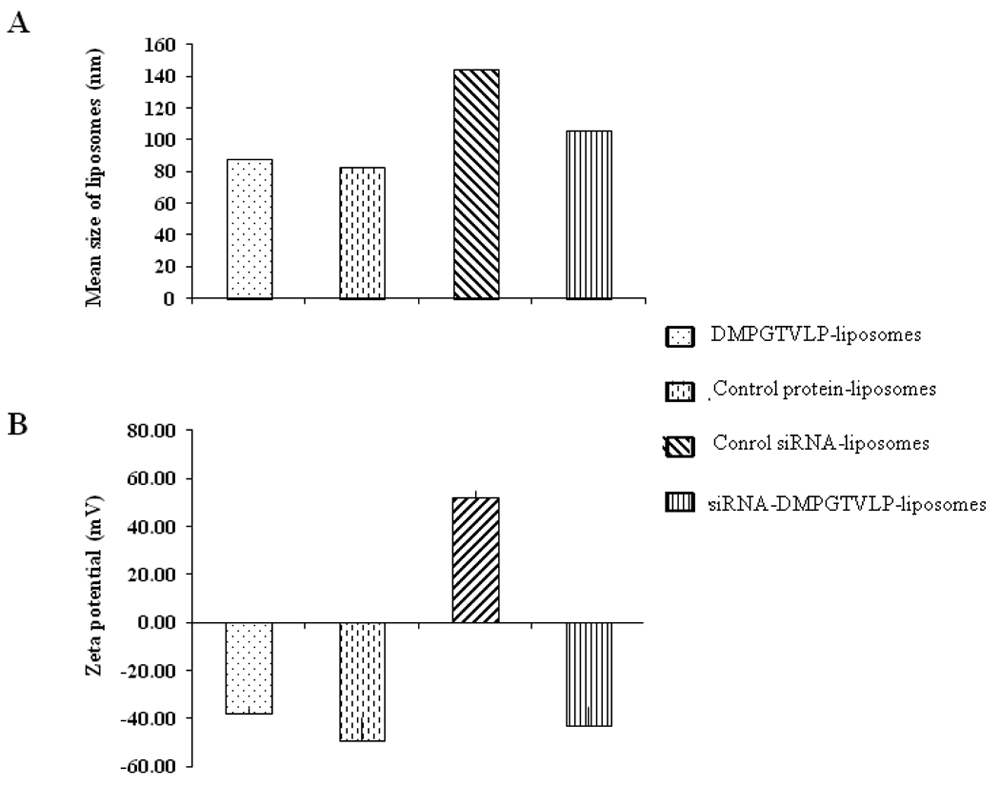
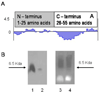
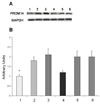
Efficacy of siRNAs as potential anticancer therapeutics can be increased by their targeted delivery into cancer cells via tumor-specific ligands. Phage display offers a unique approach to identify highly specific and selective ligands that can deliver nanocarriers to the site of disease. In this study, we proved a novel approach for intracellular delivery of siRNAs into breast cancer cells through their encapsulation into liposomes targeted to the tumor cells with preselected intact phage proteins. The targeted siRNA liposomes were obtained by a fusion of two parental liposomes containing spontaneously inserted siRNA and fusion phage proteins. The presence of pVIII coat protein fused to a MCF-7 cell-targeting peptide DMPGTVLP in the liposomes was confirmed by Western blotting. The novel phage-targeted siRNA-nanopharmaceuticals demonstrate significant down-regulation of PRDM14 gene expression and PRDM14 protein synthesis in the target MCF-7 cells. This approach offers the potential for development of new anticancer siRNA-based targeted nanomedicines.
From the clinical editor: In this study, the authors report a novel approach for targeted intracellular delivery of siRNAs into breast cancer cells through encapsulation into liposomes targeted to the tumor cells with preselected intact phage proteins.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
There are no disclosures or any conflicts of interest with regard to this publication.
Figures



References
-
- Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6:556–568. - PubMed
-
- Zheng X, Vladau C, Zhang X, Suzuki M, Ichim TE, Zhang ZX, et al. A novel in vivo siRNA delivery system specifically targeting dendritic cells and silencing CD40 genes for immunomodulation. Blood. 2009;113:2646–2654. - PubMed
-
- Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. - PubMed
-
- Wang W, Tang N, Zhang CL, Liu XJ, Hu H, Zhang ZX, et al. Cell penetrating peptides enhance intracellular translocation and function of siRNA encapsulated in Pegylated liposomes. Yao Xue Xue Bao. 2006;41:142–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

