P2CS: a database of prokaryotic two-component systems
- PMID: 21051349
- PMCID: PMC3013651
- DOI: 10.1093/nar/gkq1023
P2CS: a database of prokaryotic two-component systems
Abstract
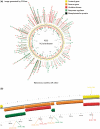
P2CS (http://www.p2cs.org) is a specialized database for prokaryotic two-component systems (TCSs), virtually ubiquitous signalling proteins which regulate a wide range of physiological processes. The primary aim of the database is to annotate and classify TCS proteins from completely sequenced prokaryotic genomes and metagenomes. Information within P2CS can be accessed through a variety of routes-TCS complements can be browsed by metagenome, replicon or sequence cluster (and these genesets are available for download by users). Alternatively a variety of database-wide or taxon-specific searches are supported. Each TCS protein is fully annotated with sequence-feature information including replicon context, while properties of the predicted proteins can be queried against several external prediction servers to suggest homologues, interaction networks, sub-cellular localization and domain complements. Another unique feature of P2CS is the analysis of ORFeomes to identify TCS genes missed during genome annotation. Recent innovations for P2CS include a CGView representation of the distribution of TCS genes around a replicon, categorization of TCS genes based on gene organization, an expanded domain-based classification scheme, a P2CS 'gene cart' and categorization on the basis of sequence clusters.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

