The mechanism for activation of GTP hydrolysis on the ribosome
- PMID: 21051640
- PMCID: PMC3763471
- DOI: 10.1126/science.1194460
The mechanism for activation of GTP hydrolysis on the ribosome
Abstract
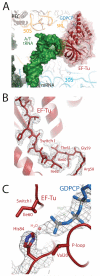
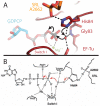
Protein synthesis requires several guanosine triphosphatase (GTPase) factors, including elongation factor Tu (EF-Tu), which delivers aminoacyl-transfer RNAs (tRNAs) to the ribosome. To understand how the ribosome triggers GTP hydrolysis in translational GTPases, we have determined the crystal structure of EF-Tu and aminoacyl-tRNA bound to the ribosome with a GTP analog, to 3.2 angstrom resolution. EF-Tu is in its active conformation, the switch I loop is ordered, and the catalytic histidine is coordinating the nucleophilic water in position for inline attack on the γ-phosphate of GTP. This activated conformation is due to a critical and conserved interaction of the histidine with A2662 of the sarcin-ricin loop of the 23S ribosomal RNA. The structure suggests a universal mechanism for GTPase activation and hydrolysis in translational GTPases on the ribosome.
Figures
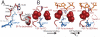
Comment in
-
Comment on "The mechanism for activation of GTP hydrolysis on the ribosome".Science. 2011 Jul 1;333(6038):37; author reply 37. doi: 10.1126/science.1202532. Science. 2011. PMID: 21719661
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

