Dating primate divergences through an integrated analysis of palaeontological and molecular data
- PMID: 21051775
- PMCID: PMC2997628
- DOI: 10.1093/sysbio/syq054
Dating primate divergences through an integrated analysis of palaeontological and molecular data
Abstract
Estimation of divergence times is usually done using either the fossil record or sequence data from modern species. We provide an integrated analysis of palaeontological and molecular data to give estimates of primate divergence times that utilize both sources of information. The number of preserved primate species discovered in the fossil record, along with their geological age distribution, is combined with the number of extant primate species to provide initial estimates of the primate and anthropoid divergence times. This is done by using a stochastic forwards-modeling approach where speciation and fossil preservation and discovery are simulated forward in time. We use the posterior distribution from the fossil analysis as a prior distribution on node ages in a molecular analysis. Sequence data from two genomic regions (CFTR on human chromosome 7 and the CYP7A1 region on chromosome 8) from 15 primate species are used with the birth-death model implemented in mcmctree in PAML to infer the posterior distribution of the ages of 14 nodes in the primate tree. We find that these age estimates are older than previously reported dates for all but one of these nodes. To perform the inference, a new approximate Bayesian computation (ABC) algorithm is introduced, where the structure of the model can be exploited in an ABC-within-Gibbs algorithm to provide a more efficient analysis.
Figures
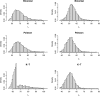
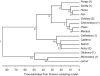
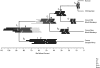
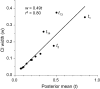
Similar articles
-
Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds.Mol Biol Evol. 2006 Jan;23(1):212-26. doi: 10.1093/molbev/msj024. Epub 2005 Sep 21. Mol Biol Evol. 2006. PMID: 16177230
-
Comparison of different strategies for using fossil calibrations to generate the time prior in Bayesian molecular clock dating.Mol Phylogenet Evol. 2017 Sep;114:386-400. doi: 10.1016/j.ympev.2017.07.005. Epub 2017 Jul 11. Mol Phylogenet Evol. 2017. PMID: 28709986 Free PMC article.
-
Estimating primate divergence times by using conditioned birth-and-death processes.Theor Popul Biol. 2009 Jun;75(4):278-85. doi: 10.1016/j.tpb.2009.02.003. Epub 2009 Mar 6. Theor Popul Biol. 2009. PMID: 19269300
-
The evolution of methods for establishing evolutionary timescales.Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20160020. doi: 10.1098/rstb.2016.0020. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325838 Free PMC article. Review.
-
Rocks and clocks revised: New promises and challenges in dating the primate tree of life.Evol Anthropol. 2022 May;31(3):138-153. doi: 10.1002/evan.21940. Epub 2022 Jan 31. Evol Anthropol. 2022. PMID: 35102633 Review.
Cited by
-
Evidence for a convergent slowdown in primate molecular rates and its implications for the timing of early primate evolution.Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6006-11. doi: 10.1073/pnas.1119506109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474376 Free PMC article.
-
Morphometric analysis of cranial shape in fossil and recent euprimates.Anat Res Int. 2012;2012:478903. doi: 10.1155/2012/478903. Epub 2012 May 7. Anat Res Int. 2012. PMID: 22611497 Free PMC article.
-
An improved assembly of the "Cascade" hop (Humulus lupulus) genome uncovers signatures of molecular evolution and refines time of divergence estimates for the Cannabaceae family.Hortic Res. 2022 Dec 7;10(2):uhac281. doi: 10.1093/hr/uhac281. eCollection 2023 Feb. Hortic Res. 2022. PMID: 36818366 Free PMC article.
-
Calibration uncertainty in molecular dating analyses: there is no substitute for the prior evaluation of time priors.Proc Biol Sci. 2015 Jan 7;282(1798):20141013. doi: 10.1098/rspb.2014.1013. Proc Biol Sci. 2015. PMID: 25429012 Free PMC article.
-
Computational Performance and Statistical Accuracy of *BEAST and Comparisons with Other Methods.Syst Biol. 2016 May;65(3):381-96. doi: 10.1093/sysbio/syv118. Epub 2016 Jan 28. Syst Biol. 2016. PMID: 26821913 Free PMC article.
References
-
- Alroy J. Chicago. University of Chicago; 1994. Quantitative mammalian biochronology and biogeography of North America [PhD thesis] (IL)
-
- Andrews P, Harrison T, Delson E, Bernor RL, Martin L. In: Distribution and biochronology of European and Southwest Asian Miocene catarrhines. The evolution of western Eurasian Neogene mammal faunas. Bernor RL, Fahlbusch V, Mittmann H-W, editors. New York: Columbia University Press; 1996. pp. 168–207.
-
- Andrews P, Kelley J. Middle Miocene dispersals of apes. Folia Primatol. 2007;78:328–343. - PubMed
-
- Azzalini A. Padova, Italy: Università di Padova; 2010. R package sn: the skew-normal and skew- tdistributions. Version 0.4-15.
-
- Azzalini A, Genton M. Robust likelihood methods based on the skew-t and related distributions. Int. Stat. Rev. 2007;76:106–129.

