Human stem cell cultures from cleft lip/palate patients show enrichment of transcripts involved in extracellular matrix modeling by comparison to controls
- PMID: 21052871
- PMCID: PMC3073041
- DOI: 10.1007/s12015-010-9197-3
Human stem cell cultures from cleft lip/palate patients show enrichment of transcripts involved in extracellular matrix modeling by comparison to controls
Abstract
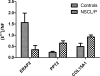
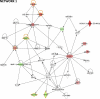
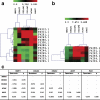
Nonsyndromic cleft lip and palate (NSCL/P) is a complex disease resulting from failure of fusion of facial primordia, a complex developmental process that includes the epithelial-mesenchymal transition (EMT). Detection of differential gene transcription between NSCL/P patients and control individuals offers an interesting alternative for investigating pathways involved in disease manifestation. Here we compared the transcriptome of 6 dental pulp stem cell (DPSC) cultures from NSCL/P patients and 6 controls. Eighty-seven differentially expressed genes (DEGs) were identified. The most significant putative gene network comprised 13 out of 87 DEGs of which 8 encode extracellular proteins: ACAN, COL4A1, COL4A2, GDF15, IGF2, MMP1, MMP3 and PDGFa. Through clustering analyses we also observed that MMP3, ACAN, COL4A1 and COL4A2 exhibit co-regulated expression. Interestingly, it is known that MMP3 cleavages a wide range of extracellular proteins, including the collagens IV, V, IX, X, proteoglycans, fibronectin and laminin. It is also capable of activating other MMPs. Moreover, MMP3 had previously been associated with NSCL/P. The same general pattern was observed in a further sample, confirming involvement of synchronized gene expression patterns which differed between NSCL/P patients and controls. These results show the robustness of our methodology for the detection of differentially expressed genes using the RankProd method. In conclusion, DPSCs from NSCL/P patients exhibit gene expression signatures involving genes associated with mechanisms of extracellular matrix modeling and palate EMT processes which differ from those observed in controls. This comparative approach should lead to a more rapid identification of gene networks predisposing to this complex malformation syndrome than conventional gene mapping technologies.
Figures

Similar articles
-
Gene expression profiling analysis contributes to understanding the association between non-syndromic cleft lip and palate, and cancer.Mol Med Rep. 2016 Mar;13(3):2110-6. doi: 10.3892/mmr.2016.4802. Epub 2016 Jan 20. Mol Med Rep. 2016. PMID: 26795696 Free PMC article.
-
Association of single nucleotide polymorphisms at 20q12 with nonsyndromic cleft lip with or without cleft palate in a Southern Chinese Han cohort.Mol Genet Genomic Med. 2020 Jan;8(1):e1028. doi: 10.1002/mgg3.1028. Epub 2019 Nov 11. Mol Genet Genomic Med. 2020. PMID: 31713353 Free PMC article.
-
Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort.Gene. 2016 Aug 15;588(1):69-73. doi: 10.1016/j.gene.2016.04.045. Epub 2016 Apr 26. Gene. 2016. PMID: 27129939
-
Association of the WNT3 polymorphisms and non-syndromic cleft lip with or without cleft palate: evidence from a meta-analysis.Biosci Rep. 2018 Nov 23;38(6):BSR20181676. doi: 10.1042/BSR20181676. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30355643 Free PMC article.
-
Polymorphic Variants of V-Maf Musculoaponeurotic Fibrosarcoma Oncogene Homolog B (rs13041247 and rs11696257) and Risk of Non-Syndromic Cleft Lip/Palate: Systematic Review and Meta-Analysis.Int J Environ Res Public Health. 2019 Aug 5;16(15):2792. doi: 10.3390/ijerph16152792. Int J Environ Res Public Health. 2019. PMID: 31387249 Free PMC article.
Cited by
-
Network-based identification of critical regulators as putative drivers of human cleft lip.BMC Med Genomics. 2019 Jan 31;12(Suppl 1):16. doi: 10.1186/s12920-018-0458-3. BMC Med Genomics. 2019. PMID: 30704473 Free PMC article.
-
E-cadherin variants associated with oral facial clefts trigger aberrant cell motility in a REG1A-dependent manner.Cell Commun Signal. 2024 Feb 27;22(1):152. doi: 10.1186/s12964-024-01532-x. Cell Commun Signal. 2024. PMID: 38414029 Free PMC article.
-
Functional impact of multi-omic interactions in breast cancer subtypes.Front Genet. 2023 Jan 5;13:1078609. doi: 10.3389/fgene.2022.1078609. eCollection 2022. Front Genet. 2023. PMID: 36685900 Free PMC article.
-
Evolutionary dynamism of the primate LRRC37 gene family.Genome Res. 2013 Jan;23(1):46-59. doi: 10.1101/gr.138842.112. Epub 2012 Oct 11. Genome Res. 2013. PMID: 23064749 Free PMC article.
-
Dentin dysplasia type I-A dental disease with genetic heterogeneity.Oral Dis. 2019 Mar;25(2):439-446. doi: 10.1111/odi.12861. Epub 2018 Apr 10. Oral Dis. 2019. PMID: 29575674 Free PMC article. Review.
References
-
- Gundlach KKH, Maus C. Epidemiological studies on the frequency of clefts in Europe and world-wide. Journal of Cranio-Maxillo-Facial Surgery. 2006;34(2):1–2. - PubMed
-
- Kerrigan JJ, Mansell JP, Sengupta A, Brown N, Sandy JR. Palatogenesis and potential mechanisms for clefting. Journal of the Royal College of Surgeons of Edinburgh. 2000;45(6):351–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

