A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla)
- PMID: 21054831
- PMCID: PMC2989969
- DOI: 10.1186/1743-422X-7-303
A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla)
Abstract
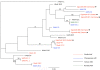
Adenoviruses (AdV) broadly infect vertebrate hosts including a variety of primates. We identified a novel AdV in the feces of captive gorillas by isolation in cell culture, electron microscopy and PCR. From the supernatants of infected cultures we amplified DNA polymerase (DPOL), preterminal protein (pTP) and hexon gene sequences with generic pan primate AdV PCR assays. The sequences in-between were amplified by long-distance PCRs of 2-10 kb length, resulting in a final sequence of 15.6 kb. Phylogenetic analysis placed the novel gorilla AdV into a cluster of primate AdVs belonging to the species Human adenovirus B (HAdV-B). Depending on the analyzed gene, its position within the cluster was variable. To further elucidate its origin, feces samples of wild gorillas were analyzed. AdV hexon sequences were detected which are indicative for three distinct and novel gorilla HAdV-B viruses, among them a virus nearly identical to the novel AdV isolated from captive gorillas. This shows that the discovered virus is a member of a group of HAdV-B viruses that naturally infect gorillas. The mixed phylogenetic clusters of gorilla, chimpanzee, bonobo and human AdVs within the HAdV-B species indicate that host switches may have been a component of the evolution of human and non-human primate HAdV-B viruses.
Figures


Similar articles
-
Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions.J Virol. 2011 Oct;85(20):10774-84. doi: 10.1128/JVI.00810-11. Epub 2011 Aug 10. J Virol. 2011. PMID: 21835802 Free PMC article.
-
Adenoviruses isolated from wild gorillas are closely related to human species C viruses.Virology. 2013 Sep;444(1-2):119-23. doi: 10.1016/j.virol.2013.05.041. Epub 2013 Jun 24. Virology. 2013. PMID: 23806387
-
Adenovirus Infections in African Humans and Wild Non-Human Primates: Great Diversity and Cross-Species Transmission.Viruses. 2020 Jun 18;12(6):657. doi: 10.3390/v12060657. Viruses. 2020. PMID: 32570742 Free PMC article.
-
Adenoviruses across the animal kingdom: a walk in the zoo.FEBS Lett. 2019 Dec;593(24):3660-3673. doi: 10.1002/1873-3468.13687. Epub 2019 Dec 9. FEBS Lett. 2019. PMID: 31747467 Review.
-
Adenovirus.Curr Top Microbiol Immunol. 2010;343:195-224. doi: 10.1007/82_2010_16. Curr Top Microbiol Immunol. 2010. PMID: 20376613 Free PMC article. Review.
Cited by
-
Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents?Int J Mol Sci. 2020 Jul 8;21(14):4821. doi: 10.3390/ijms21144821. Int J Mol Sci. 2020. PMID: 32650405 Free PMC article. Review.
-
Detection and prevalence of adenoviruses from free-ranging black howler monkeys (Alouatta pigra).Virus Genes. 2018 Dec;54(6):818-822. doi: 10.1007/s11262-018-1600-1. Epub 2018 Sep 20. Virus Genes. 2018. PMID: 30238313
-
Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions.J Virol. 2011 Oct;85(20):10774-84. doi: 10.1128/JVI.00810-11. Epub 2011 Aug 10. J Virol. 2011. PMID: 21835802 Free PMC article.
-
A first report of non-invasive adenovirus detection in wild Assamese macaques in Thailand.Primates. 2017 Apr;58(2):307-313. doi: 10.1007/s10329-016-0587-2. Epub 2016 Nov 17. Primates. 2017. PMID: 27858173
-
Multiple Cross-Species Transmission Events of Human Adenoviruses (HAdV) during Hominine Evolution.Mol Biol Evol. 2015 Aug;32(8):2072-84. doi: 10.1093/molbev/msv090. Epub 2015 Apr 9. Mol Biol Evol. 2015. PMID: 25862141 Free PMC article.
References
-
- Benkö M, Harrach B, Both G, Russel W, Adair BM, Adam E, de Jong JC, Hess M, Johnson M, Kajon A, In: Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. Elsevier; 2005. Family Adenoviridae; pp. 213–228.
-
- Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. Journal of Virology. 2009;83:8980–8985. doi: 10.1128/JVI.00786-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

