Misfolding of CasBrE SU is reversed by interactions with 4070A Env: implications for gammaretroviral neuropathogenesis
- PMID: 21054857
- PMCID: PMC2998453
- DOI: 10.1186/1742-4690-7-93
Misfolding of CasBrE SU is reversed by interactions with 4070A Env: implications for gammaretroviral neuropathogenesis
Abstract
Background: CasBrE is a neurovirulent murine leukemia virus (MLV) capable of inducing paralytic disease with associated spongiform neurodegeneration. The neurovirulence of this virus has been genetically mapped to the surface expressed subunit (SU) of the env gene. However, CasBrE SU synthesized in the absence of the transmembrane subunit (TM) does not retain ecotropic receptor binding activity, indicating that folding of the receptor binding domain (RBD) requires this domain. Using a neural stem cell (NSC) based viral trans complementation approach to examine whether misfolded CasBrE SU retained neurovirulence, we observed CasBrE SU interaction with the "non-neurovirulent" amphotropic helper virus, 4070A which restored functional activity of CasBrE SU.
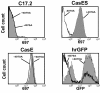
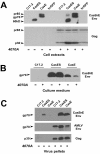
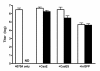
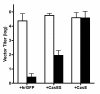
Results: Herein, we show that infection of NSCs expressing CasBrE SU with 4070A (CasES+4070A-NSCs) resulted in the redistribution of CasBrE SU from a strictly secreted product to include retention on the plasma membrane. Cell surface cross-linking analysis suggested that CasBrE SU membrane localization was due to interactions with 4070A Env. Viral particles produced from CasES+4070A-NSCS contained both CasBrE and 4070A gp70 Env proteins. These particles displayed ecotropic receptor-mediated infection, but were still 100-fold less efficient than CasE+4070A-NSC virus. Infectious center analysis showed CasBrE SU ecotropic transduction efficiencies approaching those of NSCs expressing full length CasBrE Env (CasE; SU+TM). In addition, CasBrE SU-4070A Env interactions resulted in robust ecotropic superinfection interference indicating near native intracellular SU interaction with its receptor, mCAT-1.
Conclusions: In this report we provided evidence that 4070A Env and CasBrE SU physically interact within NSCs leading to CasBrE SU retention on the plasma membrane, incorporation into viral particles, restoration of mCAT-1 binding, and capacity for initiation of TM-mediated fusion events. Thus, heterotropic Env-SU interactions facilitates CasBrE SU folding events that restore Env activity. These findings are consistent with the idea that one protein conformation acts as a folding scaffold or nucleus for a second protein of similar primary structure, a process reminiscent of prion formation. The implication is that template-based protein folding may represent an inherent feature of neuropathogenic proteins that extends to retroviral Envs.
Figures



Similar articles
-
Retrovirus-induced spongiform neurodegeneration is mediated by unique central nervous system viral targeting and expression of env alone.J Virol. 2011 Mar;85(5):2060-78. doi: 10.1128/JVI.02210-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191010 Free PMC article.
-
Reexamination of amphotropic murine leukemia virus neurovirulence: neural stem cell-mediated microglial infection fails to induce acute neurodegeneration.Virology. 2002 Feb 15;293(2):262-72. doi: 10.1006/viro.2001.1299. Virology. 2002. PMID: 11886246
-
Ecotropic Murine Leukemia Virus Infection of Glial Progenitors Interferes with Oligodendrocyte Differentiation: Implications for Neurovirulence.J Virol. 2016 Jan 13;90(7):3385-99. doi: 10.1128/JVI.03156-15. J Virol. 2016. PMID: 26764005 Free PMC article.
-
Unique N-linked glycosylation of CasBrE Env influences its stability, processing, and viral infectivity but not its neurotoxicity.J Virol. 2013 Aug;87(15):8372-87. doi: 10.1128/JVI.00392-13. Epub 2013 May 22. J Virol. 2013. PMID: 23698308 Free PMC article.
-
Erythroleukaemia induction by the Friend spleen focus-forming virus.Baillieres Clin Haematol. 1995 Mar;8(1):225-47. doi: 10.1016/s0950-3536(05)80239-2. Baillieres Clin Haematol. 1995. PMID: 7663048 Review.
Cited by
-
Retrovirus-induced spongiform neurodegeneration is mediated by unique central nervous system viral targeting and expression of env alone.J Virol. 2011 Mar;85(5):2060-78. doi: 10.1128/JVI.02210-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191010 Free PMC article.
References
-
- Gardner MB. Retroviral spongiform polioencephalomyelopathy. Rev Infect Dis. 1985;7:99–110. - PubMed
-
- Gardner MB, Rasheed S, Klement V, Rongey RW, Brown JC, Dworsky R, Henderson BE. Lower motor neuron disease in wild mice caused by indigenous type C virus and search for a similar etiology in human amyotrophic lateral sclerosis. UCLA Forum Med Sci. 1976;19:217–234. - PubMed
-
- Jolicoeur P, Gravel C, Kay DG. In: Molecular Neurovirology. Roos RP, editor. Totowa, NJ: Humana Press Inc; 1992. Pathogenesis of murine spongiform myeloencephalopathy induced by a murine retrovirus; pp. 199–224.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

