Evolution of xyloglucan-related genes in green plants
- PMID: 21054875
- PMCID: PMC3087550
- DOI: 10.1186/1471-2148-10-341
Evolution of xyloglucan-related genes in green plants
Abstract
Background: The cell shape and morphology of plant tissues are intimately related to structural modifications in the primary cell wall that are associated with key processes in the regulation of cell growth and differentiation. The primary cell wall is composed mainly of cellulose immersed in a matrix of hemicellulose, pectin, lignin and some structural proteins. Xyloglucan is a hemicellulose polysaccharide present in the cell walls of all land plants (Embryophyta) and is the main hemicellulose in non-graminaceous angiosperms.
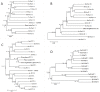
Results: In this work, we used a comparative genomic approach to obtain new insights into the evolution of the xyloglucan-related enzymatic machinery in green plants. Detailed phylogenetic analyses were done for enzymes involved in xyloglucan synthesis (xyloglucan transglycosylase/hydrolase, α-xylosidase, β-galactosidase, β-glucosidase and α-fucosidase) and mobilization/degradation (β-(1→4)-glucan synthase, α-fucosyltransferases, β-galactosyltransferases and α-xylosyl transferase) based on 12 fully sequenced genomes and expressed sequence tags from 29 species of green plants. Evidence from Chlorophyta and Streptophyta green algae indicated that part of the Embryophyta xyloglucan-related machinery evolved in an aquatic environment, before land colonization. Streptophyte algae have at least three enzymes of the xyloglucan machinery: xyloglucan transglycosylase/hydrolase, β-(1→4)-glucan synthase from the cellulose synthase-like C family and α-xylosidase that is also present in chlorophytes. Interestingly, gymnosperm sequences orthologs to xyloglucan transglycosylase/hydrolases with exclusively hydrolytic activity were also detected, suggesting that such activity must have emerged within the last common ancestor of spermatophytes. There was a positive correlation between the numbers of founder genes within each gene family and the complexity of the plant cell wall.
Conclusions: Our data support the idea that a primordial xyloglucan-like polymer emerged in streptophyte algae as a pre-adaptation that allowed plants to subsequently colonize terrestrial habitats. Our results also provide additional evidence that charophycean algae and land plants are sister groups.
Figures




References
-
- Fry SC. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annual Review of Plant Physiology, Palo Alto. 1986;37:165–186. doi: 10.1146/annurev.pp.37.060186.001121. - DOI
-
- Carpita N, McCann M. In: Biochemistry and Molecular Biology of Plants. Buchanan BB, Gruissem W, Jones RL, editor. American Society of Plant Physiologists, Rockville, Maryland; 2000. The cell wall; pp. 52–108.
-
- Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R. The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa. Plant Biol (Stuttg) 2004;6(4):402–407. doi: 10.1055/s-2004-817959. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

