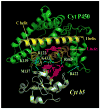
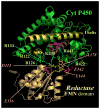
The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5)
- PMID: 21055385
- PMCID: PMC3073529
- DOI: 10.1016/j.abb.2010.10.023
The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5)
Abstract
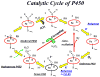
Cytochrome P450 2B4 is a microsomal protein with a multi-step reaction cycle similar to that observed in the majority of other cytochromes P450. The cytochrome P450 2B4-substrate complex is reduced from the ferric to the ferrous form by cytochrome P450 reductase. After binding oxygen, the oxyferrous protein accepts a second electron which is provided by either cytochrome P450 reductase or cytochrome b(5). In both instances, product formation occurs. When the second electron is donated by cytochrome b(5), catalysis (product formation) is ∼10- to 100-fold faster than in the presence of cytochrome P450 reductase. This allows less time for side product formation (hydrogen peroxide and superoxide) and improves by ∼15% the coupling of NADPH consumption to product formation. Cytochrome b(5) has also been shown to compete with cytochrome P450 reductase for a binding site on the proximal surface of cytochrome P450 2B4. These two different effects of cytochrome b(5) on cytochrome P450 2B4 reactivity can explain how cytochrome b(5) is able to stimulate, inhibit, or have no effect on cytochrome P450 2B4 activity. At low molar ratios (<1) of cytochrome b(5) to cytochrome P450 reductase, the more rapid catalysis results in enhanced substrate metabolism. In contrast, at high molar ratios (>1) of cytochrome b(5) to cytochrome P450 reductase, cytochrome b(5) inhibits activity by binding to the proximal surface of cytochrome P450 and preventing the reductase from reducing ferric cytochrome P450 to the ferrous protein, thereby aborting the catalytic reaction cycle. When the stimulatory and inhibitory effects of cytochrome b(5) are equal, it will appear to have no effect on the enzymatic activity. It is hypothesized that cytochrome b(5) stimulates catalysis by causing a conformational change in the active site, which allows the active oxidizing oxyferryl species of cytochrome P450 to be formed more rapidly than in the presence of reductase.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


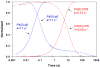
 , product formation by P450-b5; , product formation by P450-b5; |  , product formation by P450-CPR; , product formation by P450-CPR; |
 , ΔA438nm for P450-b5; , ΔA438nm for P450-b5; |  , ΔA438nm for P450-CPR. , ΔA438nm for P450-CPR. |
References
-
- Ortiz de Montellano PR. Cytochrome P450: structure, mechanism, and biochemistry. Kluwer Academic/Plenum Publishers; New York, New York: 2005.
-
- Shaik S, Cohen Y, Wang HC, Kumar D, Thiel W. P450 Enzymes: Their Structure, Reactivity and Selectivity, Modeled by QM/MM Calculations. Chem Rev. 2010;110:949. - PubMed
-
- Paine MJ, Scrutton NS, Munro AW, Gutierrez A, Roberts GCK, Wolf CR. In: Cytochrome P450. 3rd. Ortiz de Montellano PR, editor. Kluwer Academic/Plenum Publishers; New York, Boston, Dordrecht, London, Moscow: 2005. pp. 115–148.
-
- Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 2003;97:139–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

