A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability
- PMID: 21055985
- PMCID: PMC3006237
- DOI: 10.1016/j.molcel.2010.10.022
A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability
Abstract
Replication stress involving collision of replisomes with camptothecin (CPT)-stabilized DNA-Topoisomerase I adducts activates an ATR-dependent pathway to promote repair by homologous recombination. To identify human genes that protect cells from such replication stress, we performed a genome-wide CPT sensitivity screen. Among numerous candidate genes are two previously unstudied proteins: the ankyrin repeat protein NFKBIL2 and C6ORF167 (MMS22L), distantly related to yeast replication stress regulator Mms22p. MMS22L and NFKBIL2 interact with each other and with FACT (facilitator of chromatin transcription) and MCM (minichromosome maintenance) complexes. Cells depleted of NFKBIL2 or MMS22L are sensitive to DNA-damaging agents, load phosphorylated RPA onto chromatin in a CTIP-dependent manner, activate the ATR/ATRIP-CHK1 and double-strand break repair signaling pathways, and are defective in HR. This study identifies MMS22L-NFKBIL2 as components of the replication stress control pathway and provides a resource for discovery of additional components of this pathway.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


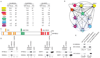
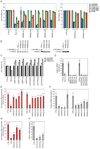


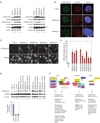
Comment in
-
Rescuing stalled replication forks: MMS22L-TONSL, a novel complex for DNA replication fork repair in human cells.Cell Cycle. 2011 Jun 1;10(11):1703-5. doi: 10.4161/cc.10.11.15557. Epub 2011 Jun 1. Cell Cycle. 2011. PMID: 21519189 No abstract available.
References
-
- Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem. 2007;282:35910–35923. - PubMed
-
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

