Transcriptional regulation of the hypocretin/orexin gene by NR6A1
- PMID: 21056546
- PMCID: PMC3012206
- DOI: 10.1016/j.bbrc.2010.11.001
Transcriptional regulation of the hypocretin/orexin gene by NR6A1
Abstract
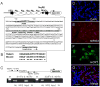
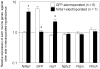
The hypocretin (also known as orexin) neuropeptide system coordinates the regulation of various physiological processes. A reduction in Nr6a1 expression was observed in hypocretin neuron-ablated transgenic mice. To show that prepro-hypocretin transcription is functionally modulated by NR6A1, we performed chromatin immunoprecipitation (ChIP) analysis, double-immunostaining, a luciferase reporter assay, and an in utero electroporation study. ChIP analysis showed that endogenous NR6A1 binds to a putative NR6A1-binding site. Double-immunostaining indicated almost all hypocretin neurons were positive for NR6A1 immunoreactivity. NR6A1 overexpression in SH-SY5Y cells modulated hypocretin promoter activity, an effect that was countered by lacking a putative NR6A1-binding site. Electroporation with Nr6a1 in the foetal hypothalamus promoted hypocretin transcription as compared to GFP-electroporation. These experiments confirmed that NR6A1 works as a regulator for hypocretin transcription.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Involvement of PLAGL1/ZAC1 in hypocretin/orexin transcription.Int J Mol Med. 2019 May;43(5):2164-2176. doi: 10.3892/ijmm.2019.4143. Epub 2019 Mar 20. Int J Mol Med. 2019. PMID: 30896835 Free PMC article.
-
The human prepro-orexin gene regulatory region that activates gene expression in the lateral region and represses it in the medial regions of the hypothalamus.J Biol Chem. 2002 May 10;277(19):16985-92. doi: 10.1074/jbc.M107962200. Epub 2002 Feb 19. J Biol Chem. 2002. PMID: 11854267
-
IGFBP3 colocalizes with and regulates hypocretin (orexin).PLoS One. 2009;4(1):e4254. doi: 10.1371/journal.pone.0004254. Epub 2009 Jan 22. PLoS One. 2009. PMID: 19158946 Free PMC article.
-
Transcriptional regulation of the hypocretin/orexin gene.Vitam Horm. 2012;89:75-90. doi: 10.1016/B978-0-12-394623-2.00005-6. Vitam Horm. 2012. PMID: 22640609 Review.
-
The hypocretin/orexin system in health and disease.Biol Psychiatry. 2003 Jul 15;54(2):87-95. doi: 10.1016/s0006-3223(03)00349-4. Biol Psychiatry. 2003. PMID: 12873797 Review.
Cited by
-
Reactivation of hyperglycemia-induced hypocretin (HCRT) gene silencing by N-acetyl-d-mannosamine in the orexin neurons derived from human iPS cells.Epigenetics. 2017 Sep;12(9):764-778. doi: 10.1080/15592294.2017.1346775. Epub 2017 Nov 24. Epigenetics. 2017. PMID: 28762874 Free PMC article.
-
Orexin Reserve: A Mechanistic Framework for the Role of Orexins (Hypocretins) in Addiction.Biol Psychiatry. 2022 Dec 1;92(11):836-844. doi: 10.1016/j.biopsych.2022.06.027. Epub 2022 Jul 3. Biol Psychiatry. 2022. PMID: 36328706 Free PMC article. Review.
-
The transcription factor HAND2 up-regulates transcription of the IL15 gene in human endometrial stromal cells.J Biol Chem. 2020 Jul 10;295(28):9596-9605. doi: 10.1074/jbc.RA120.012753. Epub 2020 May 22. J Biol Chem. 2020. PMID: 32444497 Free PMC article.
-
Hypocretin neuron-specific transcriptome profiling identifies the sleep modulator Kcnh4a.Elife. 2015 Oct 1;4:e08638. doi: 10.7554/eLife.08638. Elife. 2015. PMID: 26426478 Free PMC article.
-
Genetic manipulation of the mouse developing hypothalamus through in utero electroporation.J Vis Exp. 2013 Jul 24;(77):50412. doi: 10.3791/50412. J Vis Exp. 2013. PMID: 23912701 Free PMC article.
References
-
- de Lecea L, Sutcliffe JG. The hypocretins and sleep. Febs J. 2005;272:5675–5688. - PubMed
-
- Sakurai T. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–181. - PubMed
-
- Sakurai T, Moriguchi T, Furuya K, Kajiwara N, Nakamura T, Yanagisawa M, Goto K. Structure and function of human prepro-orexin gene. J Biol Chem. 1999;274:17771–17776. - PubMed
-
- Moriguchi T, Sakurai T, Takahashi S, Goto K, Yamamoto M. The human prepro-orexin gene regulatory region that activates gene expression in the lateral region and represses it in the medial regions of the hypothalamus. J Biol Chem. 2002;277:16985–16992. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

