Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms
- PMID: 21056574
- PMCID: PMC3060569
- DOI: 10.1016/j.jmb.2010.10.015
Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms
Abstract
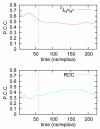
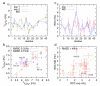
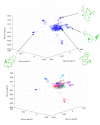
Aβ(1-42) is the highly pathologic isoform of amyloid-β, the peptide constituent of fibrils and neurotoxic oligomers involved in Alzheimer's disease. Recent studies on the structural features of Aβ in water have suggested that the system can be described as an ensemble of distinct conformational species in fast exchange. Here, we use replica exchange molecular dynamics (REMD) simulations to characterize the conformations accessible to Aβ42 in explicit water solvent, under the ff99SB force field. Monitoring the correlation between J-coupling((3)J(H(N))(H(α))) and residual dipolar coupling (RDC) data calculated from the REMD trajectories to their experimental values, as determined by NMR, indicates that the simulations converge towards sampling an ensemble that is representative of the experimental data after 60 ns/replica of simulation time. We further validate the converged MD-derived ensemble through direct comparison with (3)J(H(N))(H(α)) and RDC experimental data. Our analysis indicates that the ff99SB-derived REMD ensemble can reproduce the experimental J-coupling values with high accuracy and further provide good agreement with the RDC data. Our results indicate that the peptide is sampling a highly diverse range of conformations: by implementing statistical learning techniques (Laplacian eigenmaps, spectral clustering, and Laplacian scores), we are able to obtain an otherwise hidden structure in the complex conformational space of the peptide. Using these methods, we characterize the peptide conformations and extract their intrinsic characteristics, identify a small number of different conformations that characterize the whole ensemble, and identify a small number of protein interactions (such as contacts between the peptide termini) that are the most discriminative of the different conformations and thus can be used in designing experimental probes of transitions between such molecular states. This is a study of an important intrinsically disordered peptide system that provides an atomic-level description of structural features and interactions that are relevant during the early stages of the oligomerization and fibril nucleation pathways.
Copyright © 2010. Published by Elsevier Ltd.
Figures

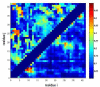
Similar articles
-
Aβ monomers transiently sample oligomer and fibril-like configurations: ensemble characterization using a combined MD/NMR approach.J Mol Biol. 2013 Sep 23;425(18):3338-59. doi: 10.1016/j.jmb.2013.06.021. Epub 2013 Jun 25. J Mol Biol. 2013. PMID: 23811057 Free PMC article.
-
Effects of force fields on the conformational and dynamic properties of amyloid β(1-40) dimer explored by replica exchange molecular dynamics simulations.Proteins. 2018 Mar;86(3):279-300. doi: 10.1002/prot.25439. Epub 2017 Dec 25. Proteins. 2018. PMID: 29235155
-
Characterization of the internal dynamics and conformational space of zinc-bound amyloid β peptides by replica-exchange molecular dynamics simulations.Eur Biophys J. 2013 Jul;42(7):575-86. doi: 10.1007/s00249-013-0906-0. Epub 2013 May 3. Eur Biophys J. 2013. PMID: 23640306
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
Cited by
-
Fluorescence-Labeled Amyloid Beta Monomer: A Molecular Dynamical Study.Molecules. 2020 Aug 1;25(15):3524. doi: 10.3390/molecules25153524. Molecules. 2020. PMID: 32752239 Free PMC article.
-
Ligand clouds around protein clouds: a scenario of ligand binding with intrinsically disordered proteins.PLoS Comput Biol. 2013;9(10):e1003249. doi: 10.1371/journal.pcbi.1003249. Epub 2013 Oct 3. PLoS Comput Biol. 2013. PMID: 24098099 Free PMC article.
-
Looking at the Disordered Proteins through the Computational Microscope.ACS Cent Sci. 2018 May 23;4(5):534-542. doi: 10.1021/acscentsci.7b00626. Epub 2018 Mar 22. ACS Cent Sci. 2018. PMID: 29805999 Free PMC article. Review.
-
Varied Probability of Staying Collapsed/Extended at the Conformational Equilibrium of Monomeric Aβ40 and Aβ42.Sci Rep. 2015 Jun 5;5:11024. doi: 10.1038/srep11024. Sci Rep. 2015. PMID: 26046578 Free PMC article.
-
Effects of different force fields and temperatures on the structural character of Abeta (12-28) peptide in aqueous solution.Int J Mol Sci. 2011;12(11):8259-74. doi: 10.3390/ijms12118259. Epub 2011 Nov 21. Int J Mol Sci. 2011. PMID: 22174662 Free PMC article.
References
-
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. - PubMed
-
- Rauk A. The chemistry of Alzheimer’s disease. Chemical Society Reviews. 2009;38:2698–2715. - PubMed
-
- Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–7. - PubMed
-
- Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–8. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

