Her2-specific multivalent adapters confer designed tropism to adenovirus for gene targeting
- PMID: 21056576
- PMCID: PMC3149894
- DOI: 10.1016/j.jmb.2010.10.040
Her2-specific multivalent adapters confer designed tropism to adenovirus for gene targeting
Abstract
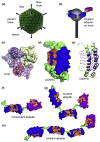
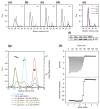
Adenoviruses (Ads) hold great promise as gene vectors for diagnostic or therapeutic applications. The native tropism of Ads must be modified to achieve disease site-specific gene delivery by Ad vectors and this should be done in a programmable way and with technology that can realistically be scaled up. To this end, we applied the technologies of designed ankyrin repeat proteins (DARPins) and ribosome display to develop a DARPin that binds the knob domain of the Ad fiber protein with low nanomolar affinity (K(D) 1.35 nM) and fused this protein with a DARPin specific for Her2, an established cell-surface biomarker of human cancers. The stability of the complex formed by this bispecific targeting adapter and the Ad virion resulted in insufficient gene transfer and was subsequently improved by increasing the valency of adapter-virus binding. In particular, we designed adapters that chelated the knob in a bivalent or trivalent fashion and showed that the efficacy of gene transfer by the adapter-Ad complex increased with the functional affinity of these molecules. This enabled efficient transduction at low stoichiometric adapter-to-fiber ratios. We confirmed the Her2 specificity of this transduction and its dependence on the Her2-binding DARPin component of the adapters. Even the adapter molecules with four fused DARPins could be produced and purified from Escherichia coli at very high levels. In principle, DARPins can be generated against any target and this adapter approach provides a versatile strategy for developing a broad range of disease-specific gene vectors.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures





Similar articles
-
Development of a generic adenovirus delivery system based on structure-guided design of bispecific trimeric DARPin adapters.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E869-77. doi: 10.1073/pnas.1213653110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431166 Free PMC article.
-
Adenovirus targeting to c-erbB-2 oncoprotein by single-chain antibody fused to trimeric form of adenovirus receptor ectodomain.Cancer Res. 2002 Jan 15;62(2):609-16. Cancer Res. 2002. PMID: 11809717
-
Generation of fiber-modified adenovirus vectors containing heterologous peptides in both the HI loop and C terminus of the fiber knob.J Gene Med. 2003 Apr;5(4):267-76. doi: 10.1002/jgm.348. J Gene Med. 2003. PMID: 12692861
-
Tropism and transduction of oncolytic adenovirus 5 vectors in cancer therapy: Focus on fiber chimerism and mosaicism, hexon and pIX.Virus Res. 2018 Sep 15;257:40-51. doi: 10.1016/j.virusres.2018.08.012. Epub 2018 Aug 17. Virus Res. 2018. PMID: 30125593 Review.
-
Adenovirus receptors: implications for tropism, treatment and targeting.Rev Med Virol. 2009 May;19(3):165-78. doi: 10.1002/rmv.612. Rev Med Virol. 2009. PMID: 19367611 Review.
Cited by
-
Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer.Mol Ther. 2013 Jan;21(1):109-18. doi: 10.1038/mt.2012.186. Epub 2012 Sep 11. Mol Ther. 2013. PMID: 22968478 Free PMC article.
-
Designer Oncolytic Adenovirus: Coming of Age.Cancers (Basel). 2018 Jun 14;10(6):201. doi: 10.3390/cancers10060201. Cancers (Basel). 2018. PMID: 29904022 Free PMC article. Review.
-
Development of a generic adenovirus delivery system based on structure-guided design of bispecific trimeric DARPin adapters.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E869-77. doi: 10.1073/pnas.1213653110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431166 Free PMC article.
-
The Adenovirus Vector Platform: Novel Insights into Rational Vector Design and Lessons Learned from the COVID-19 Vaccine.Viruses. 2023 Jan 11;15(1):204. doi: 10.3390/v15010204. Viruses. 2023. PMID: 36680244 Free PMC article. Review.
-
Artificial affinity proteins as ligands of immunoglobulins.Biomolecules. 2015 Jan 30;5(1):60-75. doi: 10.3390/biom5010060. Biomolecules. 2015. PMID: 25647098 Free PMC article. Review.
References
-
- Amalfitano A, Parks RJ. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr Gene Ther. 2002;2:111–133. - PubMed
-
- Barouch DH, Pau MG, Custers JH, Koudstaal W, Kostense S, Havenga MJ, et al. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J Immunol. 2004;172:6290–6297. - PubMed
-
- Kobinger GP, Feldmann H, Zhi Y, Schumer G, Gao G, Feldmann F, et al. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology. 2006;346:394–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

