The oligomeric state and arrangement of the active bacterial translocon
- PMID: 21056980
- PMCID: PMC3039378
- DOI: 10.1074/jbc.M110.175638
The oligomeric state and arrangement of the active bacterial translocon
Abstract
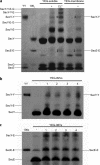
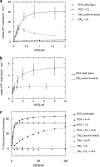
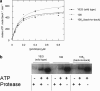
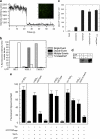
Protein secretion in bacteria is driven through the ubiquitous SecYEG complex by the ATPase SecA. The structure of SecYEG alone or as a complex with SecA in detergent reveal a monomeric heterotrimer enclosing a central protein channel, yet in membranes it is dimeric. We have addressed the functional significance of the oligomeric status of SecYEG in protein translocation using single molecule and ensemble methods. The results show that while monomers are sufficient for the SecA- and ATP-dependent association of SecYEG with pre-protein, active transport requires SecYEG dimers arranged in the back-to-back conformation. Molecular modeling of this dimeric structure, in conjunction with the new functional data, provides a rationale for the presence of both active and passive copies of SecYEG in the functional translocon.
Figures


Similar articles
-
Coassembly of SecYEG and SecA Fully Restores the Properties of the Native Translocon.J Bacteriol. 2018 Dec 7;201(1):e00493-18. doi: 10.1128/JB.00493-18. Print 2019 Jan 1. J Bacteriol. 2018. PMID: 30275279 Free PMC article.
-
Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon.J Biol Chem. 2012 Mar 9;287(11):7885-95. doi: 10.1074/jbc.M111.297911. Epub 2012 Jan 20. J Biol Chem. 2012. PMID: 22267723 Free PMC article.
-
The oligomeric distribution of SecYEG is altered by SecA and translocation ligands.J Mol Biol. 2005 Nov 25;354(2):258-71. doi: 10.1016/j.jmb.2005.09.058. Epub 2005 Oct 7. J Mol Biol. 2005. PMID: 16242710
-
Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon.Biochim Biophys Acta. 2007 Jan;1768(1):5-12. doi: 10.1016/j.bbamem.2006.08.013. Epub 2006 Aug 30. Biochim Biophys Acta. 2007. PMID: 17011510 Free PMC article. Review.
-
SecA-mediated targeting and translocation of secretory proteins.Biochim Biophys Acta. 2014 Aug;1843(8):1466-74. doi: 10.1016/j.bbamcr.2014.02.014. Epub 2014 Feb 25. Biochim Biophys Acta. 2014. PMID: 24583121 Review.
Cited by
-
Cryo-electron microscopic structure of SecA protein bound to the 70S ribosome.J Biol Chem. 2014 Mar 7;289(10):7190-7199. doi: 10.1074/jbc.M113.506634. Epub 2014 Jan 17. J Biol Chem. 2014. PMID: 24443566 Free PMC article.
-
Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex.J Cell Biol. 2012 Dec 10;199(6):919-29. doi: 10.1083/jcb.201205191. Epub 2012 Dec 3. J Cell Biol. 2012. PMID: 23209305 Free PMC article.
-
The pilus usher controls protein interactions via domain masking and is functional as an oligomer.Nat Struct Mol Biol. 2015 Jul;22(7):540-6. doi: 10.1038/nsmb.3044. Epub 2015 Jun 8. Nat Struct Mol Biol. 2015. PMID: 26052892 Free PMC article.
-
SecA, a remarkable nanomachine.Cell Mol Life Sci. 2011 Jun;68(12):2053-66. doi: 10.1007/s00018-011-0681-y. Epub 2011 Apr 10. Cell Mol Life Sci. 2011. PMID: 21479870 Free PMC article. Review.
-
A single copy of SecYEG is sufficient for preprotein translocation.EMBO J. 2011 Sep 6;30(21):4387-97. doi: 10.1038/emboj.2011.314. EMBO J. 2011. PMID: 21897368 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

