The atypical OmpR/PhoB response regulator ChxR from Chlamydia trachomatis forms homodimers in vivo and binds a direct repeat of nucleotide sequences
- PMID: 21057008
- PMCID: PMC3019824
- DOI: 10.1128/JB.00833-10
The atypical OmpR/PhoB response regulator ChxR from Chlamydia trachomatis forms homodimers in vivo and binds a direct repeat of nucleotide sequences
Abstract
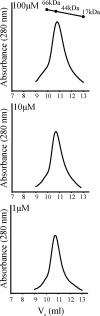
Two-component signal transduction systems are widespread in bacteria and are essential regulatory mechanisms for many biological processes. These systems predominantly rely on a sensor kinase to phosphorylate a response regulator for controlling activity, which is frequently transcriptional regulation. In recent years, an increasing number of atypical response regulators have been discovered in phylogenetically diverse bacteria. These atypical response regulators are not controlled by phosphorylation and exhibit transcriptional activity in their wild-type form. Relatively little is known regarding the mechanisms utilized by these atypical response regulators and the conserved characteristics of these atypical response regulators. Chlamydia spp. are medically important bacteria and encode an atypical OmpR/PhoB subfamily response regulator termed ChxR. In this study, protein expression analysis supports that ChxR is likely exerting its effect during the middle and late stages of the chlamydial developmental cycle, stages that include the formation of infectious elementary bodies. In the absence of detectable phosphorylation, ChxR formed homodimers in vitro and in vivo, similar to a phosphorylated OmpR/PhoB subfamily response regulator. ChxR was demonstrated to bind to its own promoter in vivo, supporting the role of ChxR as an autoactivator. Detailed analysis of the ChxR binding sites within its own promoter revealed a conserved cis-acting motif that includes a tandem repeat sequence. ChxR binds specifically to each of the individual sites and exhibits a relatively large spectrum of differential affinity. Taken together, these observations support the conclusion that ChxR, in the absence of phosphorylation, exhibits many of the characteristics of a phosphorylated (active) OmpR/PhoB subfamily response regulator.
Figures






Similar articles
-
Atypical response regulator ChxR from Chlamydia trachomatis is structurally poised for DNA binding.PLoS One. 2014 Mar 19;9(3):e91760. doi: 10.1371/journal.pone.0091760. eCollection 2014. PLoS One. 2014. PMID: 24646934 Free PMC article.
-
The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.J Biol Chem. 2011 Sep 16;286(37):32606-16. doi: 10.1074/jbc.M111.220574. Epub 2011 Jul 20. J Biol Chem. 2011. PMID: 21775428 Free PMC article.
-
Expression, purification, crystallization and preliminary X-ray analysis of the DNA-binding domain of a Chlamydia trachomatis OmpR/PhoB-subfamily response regulator homolog, ChxR.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Aug 1;65(Pt 8):791-4. doi: 10.1107/S1744309109025184. Epub 2009 Jul 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19652341 Free PMC article.
-
Prokaryotic 2-component systems and the OmpR/PhoB superfamily.Can J Microbiol. 2015 Nov;61(11):799-810. doi: 10.1139/cjm-2015-0345. Epub 2015 Aug 21. Can J Microbiol. 2015. PMID: 26382712 Review.
-
TetR and OmpR family regulators in natural product biosynthesis and resistance.Proteins. 2025 Jan;93(1):38-71. doi: 10.1002/prot.26621. Epub 2023 Oct 24. Proteins. 2025. PMID: 37874037 Review.
Cited by
-
Regulatory (pan-)genome of an obligate intracellular pathogen in the PVC superphylum.ISME J. 2016 Sep;10(9):2129-44. doi: 10.1038/ismej.2016.23. Epub 2016 Mar 8. ISME J. 2016. PMID: 26953603 Free PMC article.
-
To ∼P or Not to ∼P? Non-canonical activation by two-component response regulators.Mol Microbiol. 2017 Jan;103(2):203-213. doi: 10.1111/mmi.13532. Epub 2016 Oct 11. Mol Microbiol. 2017. PMID: 27656860 Free PMC article. Review.
-
The aspartate-less receiver (ALR) domains: distribution, structure and function.PLoS Pathog. 2015 Apr 13;11(4):e1004795. doi: 10.1371/journal.ppat.1004795. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25875291 Free PMC article.
-
Atypical response regulator ChxR from Chlamydia trachomatis is structurally poised for DNA binding.PLoS One. 2014 Mar 19;9(3):e91760. doi: 10.1371/journal.pone.0091760. eCollection 2014. PLoS One. 2014. PMID: 24646934 Free PMC article.
-
Two-component signal transduction in Corynebacterium glutamicum and other corynebacteria: on the way towards stimuli and targets.Appl Microbiol Biotechnol. 2012 Jun;94(5):1131-50. doi: 10.1007/s00253-012-4060-x. Epub 2012 Apr 28. Appl Microbiol Biotechnol. 2012. PMID: 22539022 Free PMC article. Review.
References
-
- Abdelrahman, Y. M., and R. J. Belland. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949-959. - PubMed
-
- Ainsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34:607-619. - PubMed
-
- Arribas-Bosacoma, R., S. K. Kim, C. Ferrer-Orta, A. G. Blanco, P. J. Pereira, F. X. Gomis-Ruth, B. L. Wanner, M. Coll, and M. Sola. 2007. The X-ray crystal structures of two constitutively active mutants of the Escherichia coli PhoB receiver domain give insights into activation. J. Mol. Biol. 366:626-641. - PMC - PubMed
-
- Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

