Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp
- PMID: 21057026
- PMCID: PMC3019706
- DOI: 10.1128/AEM.01272-10
Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp
Abstract
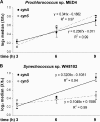
Cyanobacteria of the genera Synechococcus and Prochlorococcus are the most abundant photosynthetic organisms on earth, occupying a key position at the base of marine food webs. The cynS gene that encodes cyanase was identified among bacterial, fungal, and plant sequences in public databases, and the gene was particularly prevalent among cyanobacteria, including numerous Prochlorococcus and Synechococcus strains. Phylogenetic analysis of cynS sequences retrieved from the Global Ocean Survey database identified >60% as belonging to unicellular marine cyanobacteria, suggesting an important role for cyanase in their nitrogen metabolism. We demonstrate here that marine cyanobacteria have a functionally active cyanase, the transcriptional regulation of which varies among strains and reflects the genomic context of cynS. In Prochlorococcus sp. strain MED4, cynS was presumably transcribed as part of the cynABDS operon, implying cyanase involvement in cyanate utilization. In Synechococcus sp. strain WH8102, expression was not related to nitrogen stress responses and here cyanase presumably serves in the detoxification of cyanate resulting from intracellular urea and/or carbamoyl phosphate decomposition. Lastly, we report on a cyanase activity encoded by cynH, a novel gene found in marine cyanobacteria only. The presence of dual cyanase genes in the genomes of seven marine Synechococcus strains and their respective roles in nitrogen metabolism remain to be clarified.
Figures





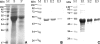
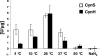

Similar articles
-
Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria.J Bacteriol. 2007 Feb;189(3):1013-24. doi: 10.1128/JB.01328-06. Epub 2006 Nov 22. J Bacteriol. 2007. PMID: 17122352 Free PMC article.
-
Characterization of the Pseudomonas pseudoalcaligenes CECT5344 Cyanase, an enzyme that is not essential for cyanide assimilation.Appl Environ Microbiol. 2008 Oct;74(20):6280-8. doi: 10.1128/AEM.00916-08. Epub 2008 Aug 15. Appl Environ Microbiol. 2008. PMID: 18708510 Free PMC article.
-
Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942.J Bacteriol. 1997 Sep;179(18):5744-50. doi: 10.1128/jb.179.18.5744-5750.1997. J Bacteriol. 1997. PMID: 9294430 Free PMC article.
-
The cyanase operon and cyanate metabolism.FEMS Microbiol Rev. 1990 Dec;7(3-4):247-52. doi: 10.1111/j.1574-6968.1990.tb04920.x. FEMS Microbiol Rev. 1990. PMID: 2094285 Review.
-
Mixotrophy in marine picocyanobacteria: use of organic compounds by Prochlorococcus and Synechococcus.ISME J. 2020 May;14(5):1065-1073. doi: 10.1038/s41396-020-0603-9. Epub 2020 Feb 7. ISME J. 2020. PMID: 32034281 Free PMC article. Review.
Cited by
-
Single cell genomic and transcriptomic evidence for the use of alternative nitrogen substrates by anammox bacteria.ISME J. 2018 Nov;12(11):2706-2722. doi: 10.1038/s41396-018-0223-9. Epub 2018 Jul 10. ISME J. 2018. PMID: 29991764 Free PMC article.
-
Satellite remote sensing data can be used to model marine microbial metabolite turnover.ISME J. 2015 Jan;9(1):166-79. doi: 10.1038/ismej.2014.107. Epub 2014 Jul 29. ISME J. 2015. PMID: 25072414 Free PMC article.
-
Isocyanic acid-mediated NLRP3 carbamoylation reduces NLRP3-NEK7 interaction and limits inflammasome activation.Sci Adv. 2025 Mar 7;11(10):eadq4266. doi: 10.1126/sciadv.adq4266. Epub 2025 Mar 7. Sci Adv. 2025. PMID: 40053593 Free PMC article.
-
Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity.ISME J. 2014 Oct;8(10):2056-68. doi: 10.1038/ismej.2014.57. Epub 2014 Apr 17. ISME J. 2014. PMID: 24739626 Free PMC article.
-
Differential global distribution of marine picocyanobacteria gene clusters reveals distinct niche-related adaptive strategies.ISME J. 2023 May;17(5):720-732. doi: 10.1038/s41396-023-01386-0. Epub 2023 Feb 25. ISME J. 2023. PMID: 36841901 Free PMC article.
References
-
- Anderson, P. M. 1980. Purification and properties of the inducible enzyme cyanase. Biochemistry 19:2882-2888. - PubMed
-
- Anderson, P. M., Y. C. Sung, and J. A. Fuchs. 1990. The cyanase operon and cyanate metabolism. FEMS Microbiol. Rev. 87:247-252. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Chin, C. C., P. M. Anderson, and F. Wold. 1983. The amino acid sequence of Escherichia coli cyanase. J. Biol. Chem. 258:276-282. - PubMed
-
- Dirnhuber, P., and F. Schütz. 1948. The isomeric transformation of urea into ammonium cyanate in aqueous solutions. Biochem. J. 42:628-632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

