Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae
- PMID: 21057056
- PMCID: PMC3019801
- DOI: 10.1128/EC.00246-10
Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae
Abstract
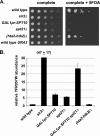
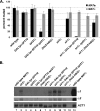
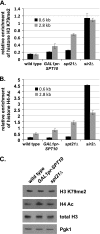
In Saccharomyces cerevisiae, transcriptional silencing occurs at three classes of genomic regions: near the telomeres, at the silent mating type loci, and within the ribosomal DNA (rDNA) repeats. In all three cases, silencing depends upon several factors, including specific types of histone modifications. In this work we have investigated the roles in silencing for Spt10 and Spt21, two proteins previously shown to control transcription of particular histone genes. Building on a recent study showing that Spt10 is required for telomeric silencing, our results show that in both spt10 and spt21 mutants, silencing is reduced near telomeres and at HMLα, while it is increased at the rDNA. Both spt10 and spt21 mutations cause modest effects on Sir protein recruitment and histone modifications at telomeric regions, and they cause significant changes in chromatin structure, as judged by its accessibility to dam methylase. These silencing and chromatin changes are not seen upon deletion of HTA2-HTB2, the primary histone locus regulated by Spt10 and Spt21. These results suggest that Spt10 and Spt21 control silencing in S. cerevisiae by altering chromatin structure through roles beyond the control of histone gene expression.
Figures





Similar articles
-
Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21.Mol Cell Biol. 2004 Jan;24(1):135-43. doi: 10.1128/MCB.24.1.135-143.2004. Mol Cell Biol. 2004. PMID: 14673149 Free PMC article.
-
SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae.Mol Cell Biol. 1994 Aug;14(8):5223-8. doi: 10.1128/mcb.14.8.5223-5228.1994. Mol Cell Biol. 1994. PMID: 8035801 Free PMC article.
-
Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1.Genetics. 2005 May;170(1):87-94. doi: 10.1534/genetics.104.039214. Epub 2005 Mar 2. Genetics. 2005. PMID: 15744051 Free PMC article.
-
SIR proteins and the assembly of silent chromatin in budding yeast.Annu Rev Genet. 2013;47:275-306. doi: 10.1146/annurev-genet-021313-173730. Epub 2013 Sep 4. Annu Rev Genet. 2013. PMID: 24016189 Review.
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
Cited by
-
Proteome-wide discovery of evolutionary conserved sequences in disordered regions.Sci Signal. 2012 Mar 13;5(215):rs1. doi: 10.1126/scisignal.2002515. Sci Signal. 2012. PMID: 22416277 Free PMC article.
-
Regulation of histone gene expression in budding yeast.Genetics. 2012 May;191(1):7-20. doi: 10.1534/genetics.112.140145. Genetics. 2012. PMID: 22555441 Free PMC article. Review.
-
Cell-cycle perturbations suppress the slow-growth defect of spt10Δ mutants in Saccharomyces cerevisiae.G3 (Bethesda). 2013 Mar;3(3):573-83. doi: 10.1534/g3.112.005389. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450643 Free PMC article.
-
A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription.PLoS Genet. 2013 May;9(5):e1003479. doi: 10.1371/journal.pgen.1003479. Epub 2013 May 2. PLoS Genet. 2013. PMID: 23658529 Free PMC article.
-
More than 10% of yeast genes are related to genome stability and influence cellular senescence via rDNA maintenance.Nucleic Acids Res. 2016 May 19;44(9):4211-21. doi: 10.1093/nar/gkw110. Epub 2016 Feb 23. Nucleic Acids Res. 2016. PMID: 26912831 Free PMC article.
References
-
- Aparicio O. M., Billington B. L., Gottschling D. E. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279–1287 - PubMed
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

