Correlated conformational events in EF-G and the ribosome regulate translocation
- PMID: 21057527
- PMCID: PMC2997181
- DOI: 10.1038/nsmb.1925
Correlated conformational events in EF-G and the ribosome regulate translocation
Abstract
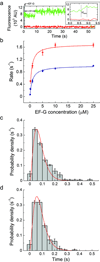
In bacteria, the translocation of tRNA and mRNA with respect to the ribosome is catalyzed by the conserved GTPase elongation factor-G (EF-G). To probe the rate-determining features in this process, we imaged EF-G-catalyzed translocation from two unique structural perspectives using single-molecule fluorescence resonance energy transfer. The data reveal that the rate at which the ribosome spontaneously achieves a transient, 'unlocked' state is closely correlated with the rate at which the tRNA-like domain IV-V element of EF-G engages the A site. After these structural transitions, translocation occurs comparatively fast, suggesting that conformational processes intrinsic to the ribosome determine the rate of translocation. Experiments conducted in the presence of non-hydrolyzable GTP analogs and specific antibiotics further reveal that allosterically linked conformational events in EF-G and the ribosome mediate rapid, directional substrate movement and EF-G release.
Figures




References
-
- Wintermeyer W, et al. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. - PubMed
-
- Fredrick K, Noller H. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. - PubMed
-
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell. 2003;12:321–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

