A DNA-binding mutant of TAL1 cooperates with LMO2 to cause T cell leukemia in mice
- PMID: 21057528
- PMCID: PMC3691994
- DOI: 10.1038/onc.2010.495
A DNA-binding mutant of TAL1 cooperates with LMO2 to cause T cell leukemia in mice
Abstract
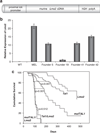
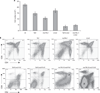
The most common translocation in childhood T-cell acute lymphoblastic leukemia (T-ALL) involves the LMO2 locus, resulting in ectopic expression of the LMO2 gene in human thymocytes. The LMO2 gene was also activated in patients with X-linked Severe Combined Immune Deficiency treated with gene therapy because of retroviral insertion in the LMO2 locus. The LMO2 insertions predisposed these children to T-ALL, yet how LMO2 contributes to T cell transformation remains unclear. The LIM (Lin 11, Isl-1, Mec-3) domain containing LMO2 protein regulates erythropoiesis as part of a large transcriptional complex consisting of LMO2, TAL1, E47, GATA1 and LDB1 that recognizes bipartite E-box-GATA1 sites on target genes. Similarly, a TAL1/E47/LMO2/LDB1 complex is observed in human T-ALL and Tal1 and Lmo2 expression in mice results in disease acceleration. To address the mechanism(s) of Tal1/Lmo2 synergy in leukemia, we generated Lmo2 transgenic mice and mated them with mice that express wild-type Tal1 or a DNA-binding mutant of TAL1. Tal1/Lmo2 and MutTAL1/Lmo2 bitransgenic mice exhibit perturbations in thymocyte development due to reduced E47/HEB transcriptional activity and develop leukemia with identical kinetics. These data demonstrate that the DNA-binding activity of Tal1 is not required to cooperate with Lmo2 to cause leukemia in mice and suggest that Lmo2 may cooperate with Tal1 to interfere with E47/HEB function(s).
© 2011 Macmillan Publishers Limited
Conflict of interest statement
The authors declare no conflict of interest.
Figures

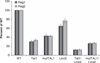

Similar articles
-
Enforced expression of E47 has differential effects on Lmo2-induced T-cell leukemias.Leuk Res. 2015 Jan;39(1):100-9. doi: 10.1016/j.leukres.2014.11.016. Epub 2014 Nov 29. Leuk Res. 2015. PMID: 25499232 Free PMC article.
-
Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex.Blood. 2011 Feb 17;117(7):2146-56. doi: 10.1182/blood-2010-07-293357. Epub 2010 Nov 12. Blood. 2011. PMID: 21076045
-
Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors.Blood. 2009 Jun 4;113(23):5783-92. doi: 10.1182/blood-2008-11-187757. Epub 2009 Jan 26. Blood. 2009. PMID: 19171877
-
Ldb1 complexes: the new master regulators of erythroid gene transcription.Trends Genet. 2014 Jan;30(1):1-9. doi: 10.1016/j.tig.2013.10.001. Epub 2013 Nov 27. Trends Genet. 2014. PMID: 24290192 Free PMC article. Review.
-
The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion.Mol Ther. 2006 Jan;13(1):15-25. doi: 10.1016/j.ymthe.2005.09.010. Epub 2005 Nov 2. Mol Ther. 2006. PMID: 16260184 Review.
Cited by
-
ESRRB regulates glucocorticoid gene expression in mice and patients with acute lymphoblastic leukemia.Blood Adv. 2020 Jul 14;4(13):3154-3168. doi: 10.1182/bloodadvances.2020001555. Blood Adv. 2020. PMID: 32658986 Free PMC article.
-
Oncogene-Induced Reprogramming in Acute Lymphoblastic Leukemia: Towards Targeted Therapy of Leukemia-Initiating Cells.Cancers (Basel). 2021 Nov 2;13(21):5511. doi: 10.3390/cancers13215511. Cancers (Basel). 2021. PMID: 34771671 Free PMC article. Review.
-
Next-Generation Sequencing in Acute Lymphoblastic Leukemia.Int J Mol Sci. 2019 Jun 15;20(12):2929. doi: 10.3390/ijms20122929. Int J Mol Sci. 2019. PMID: 31208040 Free PMC article. Review.
-
PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation.Nat Commun. 2014 May 29;5:3995. doi: 10.1038/ncomms4995. Nat Commun. 2014. PMID: 24874575 Free PMC article.
-
Whole-genome noncoding sequence analysis in T-cell acute lymphoblastic leukemia identifies oncogene enhancer mutations.Blood. 2017 Jun 15;129(24):3264-3268. doi: 10.1182/blood-2017-03-771162. Epub 2017 Apr 13. Blood. 2017. PMID: 28408461 Free PMC article.
References
-
- Abraham N, Miceli MC, Parnes JR, Veillette A. Enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Nature. 1991;350:62–66. - PubMed
-
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

