Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction
- PMID: 21059658
- PMCID: PMC3020773
- DOI: 10.1074/jbc.M110.106922
Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction
Abstract
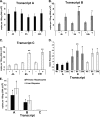
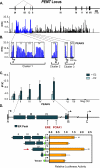
When dietary choline is restricted, most men and postmenopausal women develop multiorgan dysfunction marked by hepatic steatosis (choline deficiency syndrome (CDS)). However, a significant subset of premenopausal women is protected from CDS. Because hepatic PEMT (phosphatidylethanolamine N-methyltransferase) catalyzes de novo biosynthesis of choline and this gene is under estrogenic control, we hypothesized that there are SNPs in PEMT that disrupt the hormonal regulation of PEMT and thereby put women at risk for CDS. In this study, we performed transcript-specific gene expression analysis, which revealed that estrogen regulates PEMT in an isoform-specific fashion. Locus-wide SNP analysis identified a risk-associated haplotype that was selectively associated with loss of hormonal activation. Chromatin immunoprecipitation, analyzed by locus-wide microarray studies, comprehensively identified regions of estrogen receptor binding in PEMT. The polymorphism (rs12325817) most highly linked with the development of CDS (p < 0.00006) was located within 1 kb of the critical estrogen response element. The risk allele failed to bind either the estrogen receptor or the pioneer factor FOXA1. These data demonstrate that allele-specific ablation of estrogen receptor-DNA interaction in the PEMT locus prevents hormone-inducible PEMT expression, conferring risk of CDS in women.
Figures




References
-
- Li Z., Agellon L. B., Vance D. E. (2005) J. Biol. Chem. 280, 37798–37802 - PubMed
-
- Buchman A. L., Ament M. E., Sohel M., Dubin M., Jenden D. J., Roch M., Pownall H., Farley W., Awal M., Ahn C. (2001) JPEN J. Parenter. Enteral Nutr. 25, 260–268 - PubMed
-
- Zeisel S. H., Da Costa K. A., Franklin P. D., Alexander E. A., Lamont J. T., Sheard N. F., Beiser A. (1991) FASEB J. 5, 2093–2098 - PubMed
-
- Bremer J., Greenberg D. M. (1961) Biochim. Biophys. Acta 46, 205–216
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

